4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
8OWZ
 
 | | Crystal structure of human Sirt2 in complex with a triazole-based SirReal | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[5-[[3-[[1-(2-methoxyethyl)-1,2,3-triazol-4-yl]methoxy]phenyl]methyl]-1,3-thiazol-2-yl]ethanamide, ... | | Authors: | Friedrich, F, Zhang, L, Schiedel, M, Einsle, O, Jung, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of First-in-Class Dual Sirt2/HDAC6 Inhibitors as Molecular Tools for Dual Inhibition of Tubulin Deacetylation.
J.Med.Chem., 66, 2023
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E8J
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E9C
 
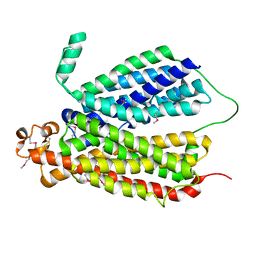 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
5ZAT
 
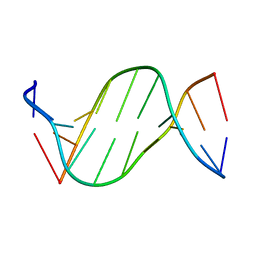 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5ZAS
 
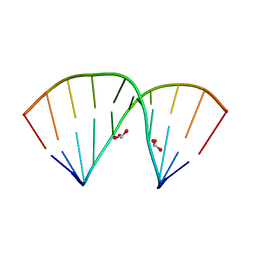 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5IDK
 
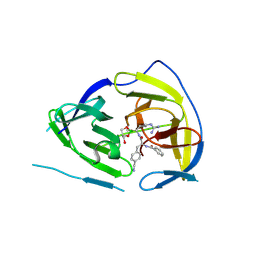 | |
5J19
 
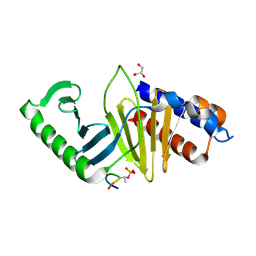 | | phospho-Pon binding-induced Plk1 dimerization | | Descriptor: | GLYCEROL, Phosphorylated peptide from Partner of Numb, Serine/threonine-protein kinase PLK1 | | Authors: | Zhu, K, Shan, Z, Zhang, L, Wen, W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phospho-Pon Binding-Mediated Fine-Tuning of Plk1 Activity
Structure, 24, 2016
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5KC2
 
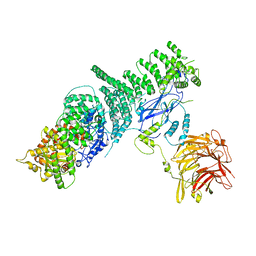 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
8IF8
 
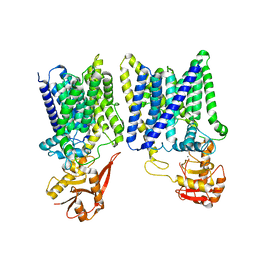 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4GWG
 
 | |
4GPZ
 
 | |
4GWK
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase complexed with 3-phosphoglyceric acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PHOSPHOGLYCERIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | He, C, Zhou, L, Zhang, L. | | Deposit date: | 2012-09-03 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth.
Cancer Cell, 22, 2012
|
|
4GXO
 
 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQM
 
 | | The crystal structure of QsrR-menadione complex | | Descriptor: | 2-methylnaphthalene-1,4-diol, QsrR protein | | Authors: | Ji, Q, Zhang, L, Jones, M.B, Sun, F, Deng, X, Liang, H, Brugarolas, P, Gao, N, Peterson, S.N, Lan, L, Bae, T, He, C. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KFP
 
 | | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived Ureas as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[1-(tetrahydro-2H-pyran-4-yl)piperidin-4-yl]sulfonyl}benzyl)-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Bair, K.W, Baumeister, T, Ho, Y, Liederer, B.M, Liu, X, O'Brien, T, Oeh, J, Sampath, D, Skelton, N, Wang, L, Wang, W, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X. | | Deposit date: | 2013-04-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived ureas as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3B8V
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221K | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
