1C1Z
 
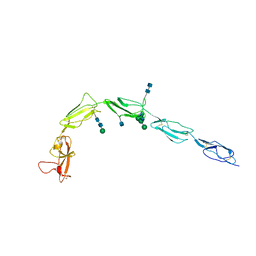 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | 著者 | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | 登録日 | 1999-07-22 | | 公開日 | 1999-11-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
2AZ1
 
 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum | | 分子名称: | CALCIUM ION, Nucleoside diphosphate kinase | | 著者 | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | 登録日 | 2005-09-09 | | 公開日 | 2005-12-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
4N58
 
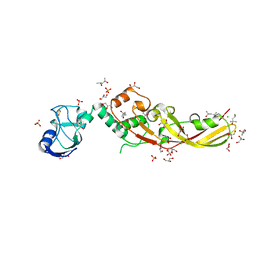 | | Crystal Structure of Pectocin M2 at 1.86 Angstroms | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Grinter, R, Roszak, A.W, Zeth, K, Cogdell, C.J, Walker, D. | | 登録日 | 2013-10-09 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4GN0
 
 | | De novo phasing of a Hamp-complex using an improved Arcimboldo method | | 分子名称: | Hamp domain of AF1503, MAGNESIUM ION | | 著者 | Hulko, M, Ursinus, A, Bar, K, Martin, J, Zeth, K, Lupas, A.N. | | 登録日 | 2012-08-16 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | 分子名称: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | 著者 | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | 登録日 | 2008-08-15 | | 公開日 | 2008-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CWO
 
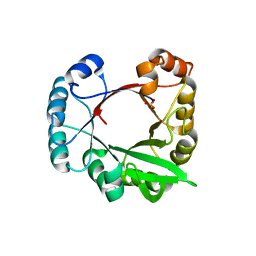 | | A beta/alpha-barrel built by the combination of fragments from different folds | | 分子名称: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | 著者 | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | 登録日 | 2008-04-22 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3FEW
 
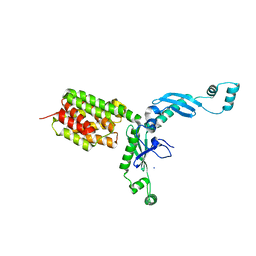 | | Structure and Function of Colicin S4, a colicin with a duplicated receptor binding domain | | 分子名称: | Colicin S4, SODIUM ION | | 著者 | Arnold, T, Linke, D, Zeth, K. | | 登録日 | 2008-12-01 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure and Function of Colicin S4, a Colicin with a Duplicated Receptor-binding Domain
J.Biol.Chem., 284, 2009
|
|
2JAF
 
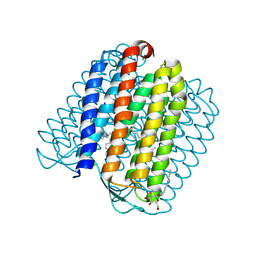 | | Ground state of halorhodopsin T203V | | 分子名称: | CHLORIDE ION, Halorhodopsin, PALMITIC ACID, ... | | 著者 | Gmelin, W, Zeth, K, Efremov, R, Heberle, J, Tittor, J, Oesterhelt, D. | | 登録日 | 2006-11-28 | | 公開日 | 2006-12-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The crystal structure of the L1 intermediate of halorhodopsin at 1.9 angstroms resolution.
Photochem. Photobiol., 83, 2007
|
|
2JAG
 
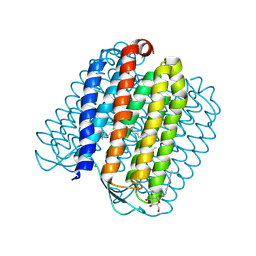 | | L1-intermediate of halorhodopsin T203V | | 分子名称: | CHLORIDE ION, Halorhodopsin, PALMITIC ACID, ... | | 著者 | Gmelin, W, Zeth, K, Efremov, R, Heberle, J, Tittor, J, Oesterhelt, D. | | 登録日 | 2006-11-28 | | 公開日 | 2006-12-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The crystal structure of the L1 intermediate of halorhodopsin at 1.9 angstroms resolution.
Photochem. Photobiol., 83, 2007
|
|
2VBV
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and FMN | | 分子名称: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | 登録日 | 2007-09-16 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBU
 
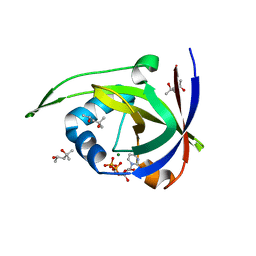 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | 登録日 | 2007-09-16 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBS
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with PO4 | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | 著者 | Hartmann, M.D, Djuranovic, S, Ammelburg, M, Martin, J, Lupas, A.N, Zeth, K. | | 登録日 | 2007-09-16 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBT
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and PO4 | | 分子名称: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | 著者 | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | 登録日 | 2007-09-16 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2V43
 
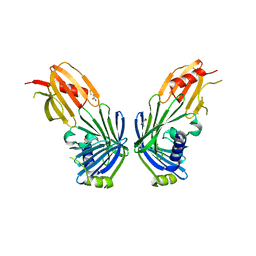 | |
2V42
 
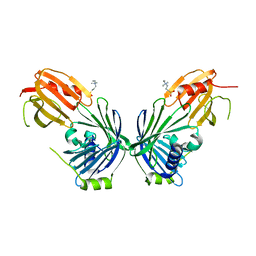 | |
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | 分子名称: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | 著者 | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | 登録日 | 2009-04-15 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2W9R
 
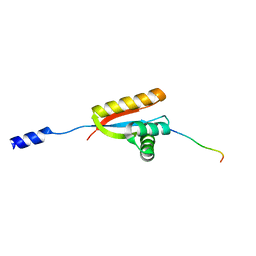 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS | | 分子名称: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, DNA PROTECTION DURING STARVATION PROTEIN | | 著者 | Schuenemann, V, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | 登録日 | 2009-01-28 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2W1T
 
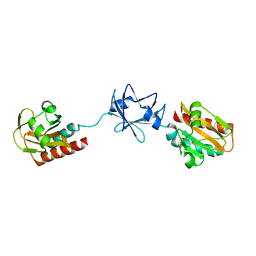 | | Crystal Structure of B. subtilis SpoVT | | 分子名称: | STAGE V SPORULATION PROTEIN T | | 著者 | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | 登録日 | 2008-10-20 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | 分子名称: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | 著者 | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | 登録日 | 2009-04-15 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WA9
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - Trp peptide structure | | 分子名称: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, TRP PEPTIDE | | 著者 | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | 登録日 | 2009-02-03 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2W1R
 
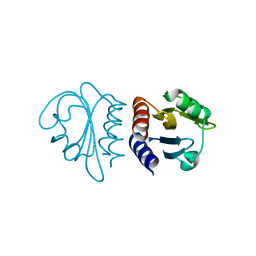 | | Crystal Structure of the C-terminal Domain of B. subtilis SpoVT | | 分子名称: | STAGE V SPORULATION PROTEIN T | | 著者 | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | 登録日 | 2008-10-20 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2WFW
 
 | | Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases - The Arc domain structure | | 分子名称: | ARC | | 著者 | Djuranovic, S, Hartmann, M.D, Habeck, M, Ursinus, A, Zwickl, P, Martin, J, Lupas, A.N, Zeth, K. | | 登録日 | 2009-04-15 | | 公開日 | 2009-05-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WA8
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - The Phe peptide structure | | 分子名称: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, N-END RULE PEPTIDE | | 著者 | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | 登録日 | 2009-02-03 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2X8X
 
 | |
2AZ3
 
 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum in complex with CDP | | 分子名称: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | 著者 | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | 登録日 | 2005-09-09 | | 公開日 | 2005-12-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
