4INK
 
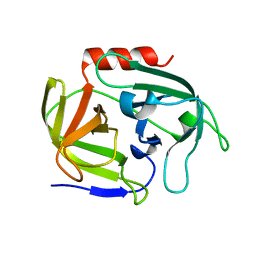 | | Crystal structure of SplD protease from Staphylococcus aureus at 1.56 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Zdzalik, M, Kalinska, M, Cichon, P, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
4MVN
 
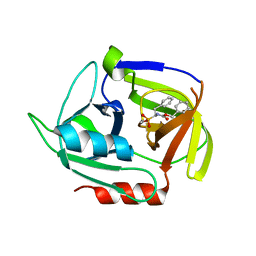 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
5N4K
 
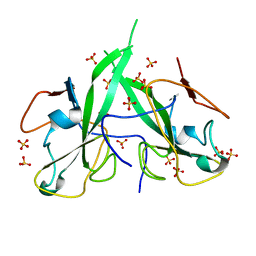 | | N-terminal domain of a human Coronavirus NL63 nucleocapsid protein | | Descriptor: | Nucleoprotein, SULFATE ION | | Authors: | Zdzalik, M, Szelazek, B, Kabala, W, Golik, P, Burmistrz, M, Florek, D, Kus, K, Pyrc, K, Dubin, G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
3UFA
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | CHLORIDE ION, N-(3-carboxypropanoyl)-L-valyl-N-[(1S)-2-phenyl-1-phosphonoethyl]-L-prolinamide, Serine protease splA | | Authors: | Zdzalik, M, Pietrusewicz, E, Pustelny, K, Stec-Niemczyk, J, Popowicz, G.M, Potempa, J, Oleksyszyn, J, Dubin, G. | | Deposit date: | 2011-10-31 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
4INL
 
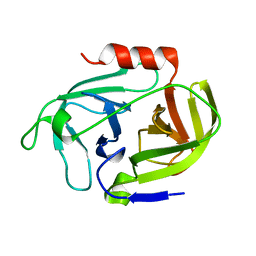 | | Crystal structure of SplD protease from Staphylococcus aureus at 2.1 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Cichon, P, Zdzalik, M, Kalinska, M, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
4K1T
 
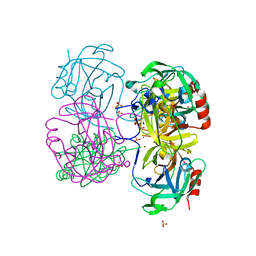 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.60 A resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine protease SplB, ... | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
4K1S
 
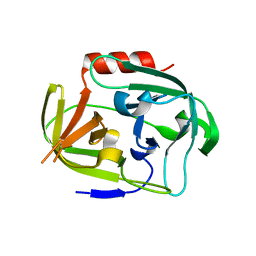 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
5MM8
 
 | |
5EPW
 
 | | C-Terminal Domain Of Human Coronavirus Nl63 Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Szelazek, B, Kabala, W, Kus, K, Zdzalik, M, Golik, P, Florek, D, Burmistrz, M, Pyrc, K, Dubin, G. | | Deposit date: | 2015-11-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
4TUJ
 
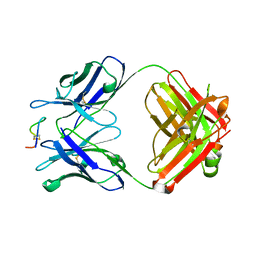 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, peptide1 | | Authors: | Grudnik, P, Golik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TRP
 
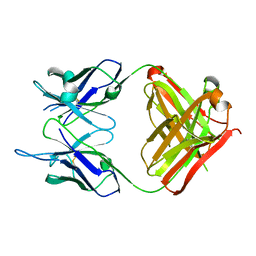 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen | | Authors: | Horwacik, I, Golik, P, Grudnik, P, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUL
 
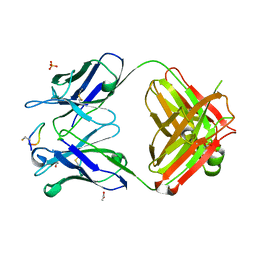 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | ACETATE ION, Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, ... | | Authors: | Golik, P, Grudnik, P, Dubin, G, Zdzalik, M, Rokita, H, Horwacik, I. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUK
 
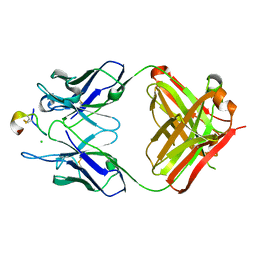 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | CHLORIDE ION, Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, ... | | Authors: | Golik, P, Grudnik, P, Dubin, G, Horwacik, I, Zdzalik, M, Rokita, H. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUO
 
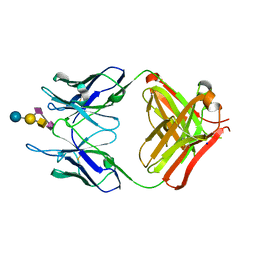 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Golik, P, Grudnik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
