1QTR
 
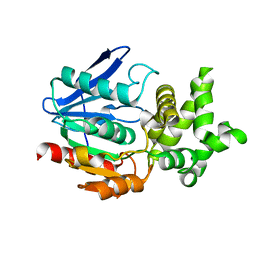 | | CRYSTAL STRUCTURE ANALYSIS OF THE PROLYL AMINOPEPTIDASE FROM SERRATIA MARCESCENS | | 分子名称: | PROLYL AMINOPEPTIDASE | | 著者 | Yoshimoto, T, Kabashima, T, Uchikawa, K, Inoue, T, Tanaka, N. | | 登録日 | 1999-06-28 | | 公開日 | 1999-07-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structure of prolyl aminopeptidase from Serratia marcescens.
J.Biochem.(Tokyo), 126, 1999
|
|
1J2T
 
 | | Creatininase Mn | | 分子名称: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-01-11 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J2U
 
 | | Creatininase Zn | | 分子名称: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-01-11 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1V7Z
 
 | | creatininase-product complex | | 分子名称: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-12-26 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1AUG
 
 | | CRYSTAL STRUCTURE OF THE PYROGLUTAMYL PEPTIDASE I FROM BACILLUS AMYLOLIQUEFACIENS | | 分子名称: | PYROGLUTAMYL PEPTIDASE-1 | | 著者 | Odagaki, Y, Hayashi, A, Okada, K, Hirotsu, K, Kabashima, T, Ito, K, Yoshimoto, T, Tsuru, D, Sato, M, Clardy, J. | | 登録日 | 1997-08-26 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of pyroglutamyl peptidase I from Bacillus amyloliquefaciens reveals a new structure for a cysteine protease.
Structure Fold.Des., 7, 1999
|
|
1KOL
 
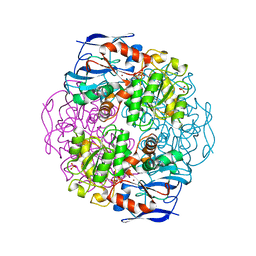 | | Crystal structure of formaldehyde dehydrogenase | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ZINC ION, ... | | 著者 | Tanaka, N, Kusakabe, Y, Ito, K, Yoshimoto, T, Nakamura, K.T. | | 登録日 | 2001-12-21 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of Formaldehyde Dehydrogenase from Pseudomonas putida: the Structural Origin of the Tightly Bound Cofactor in Nicotinoprotein Dehydrogenases
J.mol.biol., 324, 2002
|
|
1WM1
 
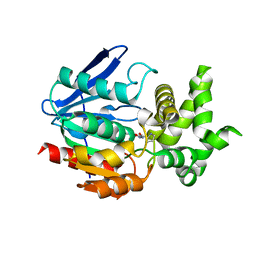 | | Crystal Structure of Prolyl Aminopeptidase, Complex with Pro-TBODA | | 分子名称: | (5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)[(2R)-PYRROLIDIN-2-YL]METHANONE, Proline iminopeptidase | | 著者 | Nakajima, Y, Inoue, T, Ito, K, Tozaka, T, Hatakeyama, S, Tanaka, N, Nakamura, K.T, Yoshimoto, T. | | 登録日 | 2004-07-01 | | 公開日 | 2004-07-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor
ARCH.BIOCHEM.BIOPHYS., 416, 2003
|
|
2D5L
 
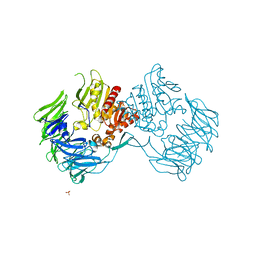 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | 分子名称: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | 著者 | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | 登録日 | 2005-11-02 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2DCM
 
 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | 分子名称: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | 著者 | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | 登録日 | 2006-01-09 | | 公開日 | 2006-09-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2ECF
 
 | |
2DQM
 
 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | 著者 | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2006-05-29 | | 公開日 | 2006-08-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2DQ6
 
 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | 分子名称: | Aminopeptidase N, SULFATE ION, ZINC ION | | 著者 | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2006-05-22 | | 公開日 | 2006-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2EEP
 
 | | Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor | | 分子名称: | Dipeptidyl aminopeptidase IV, putative, SULFATE ION, ... | | 著者 | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2007-02-16 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | 分子名称: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | 著者 | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2007-06-07 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2Z3Z
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A complexd with an inhibitor | | 分子名称: | Dipeptidyl aminopeptidase IV, SULFATE ION, [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN-2-YL]BORONIC ACID | | 著者 | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2007-06-09 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
1WMB
 
 | | Crystal structure of NAD dependent D-3-hydroxybutylate dehydrogenase | | 分子名称: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION | | 著者 | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | 登録日 | 2004-07-06 | | 公開日 | 2005-09-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
1X2E
 
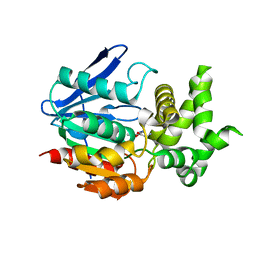 | | The crystal structure of prolyl aminopeptidase complexed with Ala-TBODA | | 分子名称: | (2S)-2-AMINO-1-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)PROPAN-1-ONE, Proline iminopeptidase | | 著者 | Nakajima, Y, Ito, K, Sakata, M, Xu, Y, Matsubara, F, Hatakeyama, S, Yoshimoto, T. | | 登録日 | 2005-04-22 | | 公開日 | 2006-05-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Unusual extra space at the active site and high activity for acetylated hydroxyproline of prolyl aminopeptidase from Serratia marcescens
J.Bacteriol., 188, 2006
|
|
1X1T
 
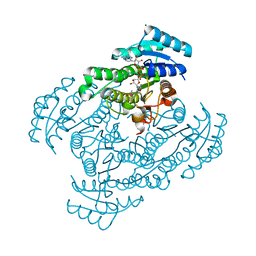 | | Crystal Structure of D-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi Complexed with NAD+ | | 分子名称: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | 著者 | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | 登録日 | 2005-04-13 | | 公開日 | 2006-01-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
1X2B
 
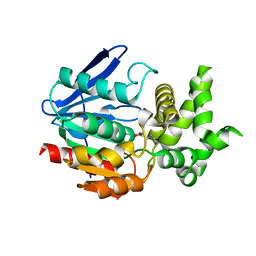 | | The crystal structure of prolyl aminopeptidase complexed with Sar-TBODA | | 分子名称: | 1-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-2-(METHYLAMINO)ETHANONE, Proline iminopeptidase | | 著者 | Nakajima, Y, Ito, K, Sakata, M, Xu, Y, Matsubara, F, Hatakeyama, S, Yoshimoto, T. | | 登録日 | 2005-04-22 | | 公開日 | 2006-05-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Unusual extra space at the active site and high activity for acetylated hydroxyproline of prolyl aminopeptidase from Serratia marcescens
J.Bacteriol., 188, 2006
|
|
3ASV
 
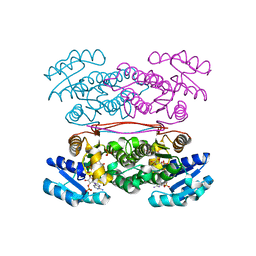 | | The Closed form of serine dehydrogenase complexed with NADP+ | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHATE ION, Short-chain dehydrogenase/reductase SDR | | 著者 | Yamazawa, R, Nakajima, Y, Yoshimoto, T, Ito, K. | | 登録日 | 2010-12-21 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of serine dehydrogenase from Escherichia coli: important role of the C-terminal region for closed-complex formation.
J.Biochem., 149, 2011
|
|
3ASU
 
 | |
3A6F
 
 | | W174F mutant creatininase, Type II | | 分子名称: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | 著者 | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | 登録日 | 2009-08-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6G
 
 | | W154F mutant creatininase | | 分子名称: | Creatinine amidohydrolase, MANGANESE (II) ION, ZINC ION | | 著者 | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | 登録日 | 2009-08-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | 分子名称: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | 著者 | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | 登録日 | 2009-08-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | 分子名称: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | 著者 | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | 登録日 | 2009-09-02 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
