6A41
 
 | | Dehalogenation enzyme | | 分子名称: | dehalogenase | | 著者 | Yin, B, Yuan, A.Y. | | 登録日 | 2018-06-18 | | 公開日 | 2019-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structure of a new dehalogenase at 1.9 Angstroms resolution
To Be Published
|
|
3WJ7
 
 | | Crystal structure of gox2253 | | 分子名称: | MERCURY (II) ION, Putative oxidoreductase | | 著者 | Yuan, Y.A, Yin, B. | | 登録日 | 2013-10-05 | | 公開日 | 2014-06-04 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans.
Proteins, 82, 2014
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | 著者 | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | 登録日 | 2018-04-12 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | 著者 | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | 登録日 | 2018-04-12 | | 公開日 | 2019-01-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3VZ1
 
 | |
3VZ2
 
 | |
3VZ3
 
 | | Structural insights into substrate and cofactor selection by sp2771 | | 分子名称: | 4-oxobutanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | 著者 | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | 登録日 | 2012-10-09 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3VZ0
 
 | | Structural insights into cofactor and substrate selection by Gox0499 | | 分子名称: | NONAETHYLENE GLYCOL, Putative NAD-dependent aldehyde dehydrogenase | | 著者 | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | 登録日 | 2012-10-09 | | 公開日 | 2013-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3WJS
 
 | |
3WUY
 
 | | Crystal structure of Nit6803 | | 分子名称: | Nitrilase | | 著者 | Yuan, Y.A, Yin, B, Wang, C. | | 登録日 | 2014-05-09 | | 公開日 | 2014-12-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
6K98
 
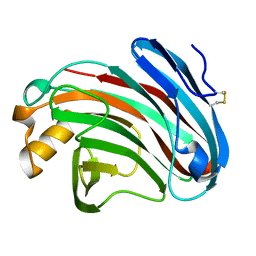 | | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family | | 分子名称: | GH12 beta-1, 4-endoglucanase | | 著者 | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, yingguo, B, Bin, Y. | | 登録日 | 2019-06-14 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.032 Å) | | 主引用文献 | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
6K9D
 
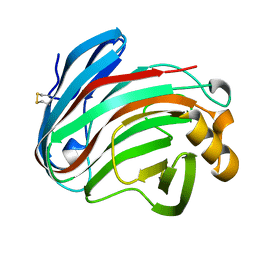 | | glycoside hydrolase family 12 (GH12) englucanase | | 分子名称: | GH12 beta-1, 4-endoglucanase | | 著者 | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, Yingguo, B, Bin, Y. | | 登録日 | 2019-06-14 | | 公開日 | 2020-06-17 | | 実験手法 | X-RAY DIFFRACTION (1.505 Å) | | 主引用文献 | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
3G36
 
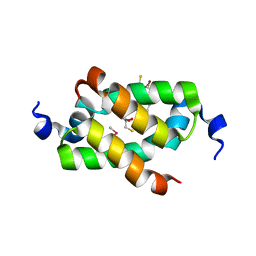 | | Crystal structure of the human DPY-30-like C-terminal domain | | 分子名称: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Wang, X, Lou, Z, Bartlam, M, Rao, Z. | | 登録日 | 2009-02-02 | | 公開日 | 2009-06-30 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of the C-terminal domain of human DPY-30-like protein: A component of the histone methyltransferase complex
J.Mol.Biol., 390, 2009
|
|
7TA6
 
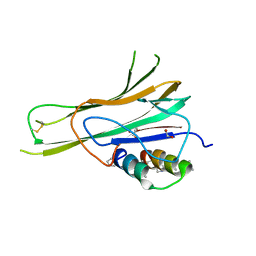 | | Trimer-to-Monomer Disruption of Tumor Necrosis Factor-alpha (TNF-alpha) by unnatural alpha/beta-peptide-1 | | 分子名称: | 1,2-ETHANEDIOL, AMINO GROUP, Alpha/Beta-peptide-1, ... | | 著者 | Niu, J, Bingman, C.A, Gellman, S.H. | | 登録日 | 2021-12-20 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Trimer-to-Monomer Disruption Mechanism for a Potent, Protease-Resistant Antagonist of Tumor Necrosis Factor-alpha Signaling.
J.Am.Chem.Soc., 144, 2022
|
|
7TA3
 
 | |
5ZBO
 
 | | Cryo-EM structure of PCV2 VLPs | | 分子名称: | Capsid protein | | 著者 | Mo, X, Yuan, A.Y. | | 登録日 | 2018-02-12 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structural roles of PCV2 capsid protein N-terminus in PCV2 particle assembly and identification of PCV2 type-specific neutralizing epitope.
PLoS Pathog., 15, 2019
|
|
5ZJU
 
 | |
7YK1
 
 | | Structural basis of human PRPS2 filaments | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Lu, G.M, Hu, H.H, Liu, J.L. | | 登録日 | 2022-07-21 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structural basis of human PRPS2 filaments.
Cell Biosci, 13, 2023
|
|
7WXI
 
 | | GPR domain of Drosophila P5CS filament with glutamate and ATPgammaS | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXH
 
 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXG
 
 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WX4
 
 | |
7WX3
 
 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXF
 
 | |
7XMV
 
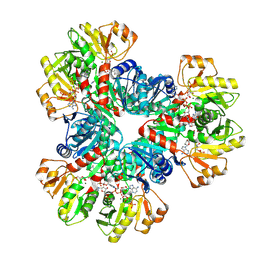 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A(AMP/ADP) filament bound with ADP, AMP and R5P | | 分子名称: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | 登録日 | 2022-04-27 | | 公開日 | 2022-06-29 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
