4XH0
 
 | | Structure of C. glabrata Hrr25 bound to ADP (SO4 condition) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-04 | | Release date: | 2016-01-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
6P80
 
 | | Structure of E. coli MS115-1 CdnC + ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Azimi, C.S, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
4XHG
 
 | | Structure of C. glabrata Hrr25 bound to ADP (formate condition) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
6P8P
 
 | | Structure of P. aeruginosa ATCC27853 HORMA1 | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8R
 
 | | Structure of P. aeruginosa ATCC27853 HORMA2 | | Descriptor: | HORMA domain containing protein, PHOSPHATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6PB3
 
 | | Structure of Rhizobiales Trip13 | | Descriptor: | Rhizobiales Sp. Pch2, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P82
 
 | | Structure of P. aeruginosa ATCC27853 CdnD, Apo form 1 | | Descriptor: | Nucleotidyltransferase, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6WZO
 
 | |
6WZQ
 
 | |
1C9W
 
 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
2F2O
 
 | |
6P8O
 
 | | Structure of P. aeruginosa ATCC27853 HORMA2-deltaC | | Descriptor: | CHLORIDE ION, HORMA domain containing protein, NICKEL (II) ION | | Authors: | Ye, Q, Corbett, K.D, Lau, R.K. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
3LMI
 
 | |
3LKM
 
 | |
3LLA
 
 | |
3LMH
 
 | |
1I71
 
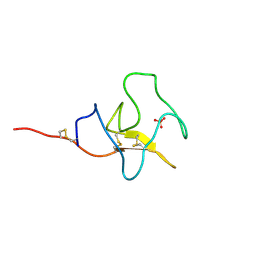 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF APOLIPOPROTEIN(A) KRINGLE IV TYPE 7: INSIGHTS INTO LIGAND BINDING | | Descriptor: | APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Ye, Q, Rahman, M.N, Koschinsky, M.L, Jia, Z. | | Deposit date: | 2001-03-07 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of apolipoprotein(a) kringle IV type 7: insights into ligand binding.
Protein Sci., 10, 2001
|
|
6XNR
 
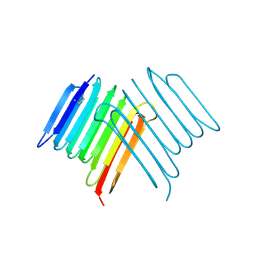 | | Crystal structure of Rhagium Mordax antifreeze protein | | Descriptor: | 1,2-ETHANEDIOL, Antifreeze protein | | Authors: | Ye, Q, Eves, R, Campbell, R.L, Davies, P.L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an insect antifreeze protein reveals ordered waters on the ice-binding surface.
Biochem.J., 477, 2020
|
|
1HQT
 
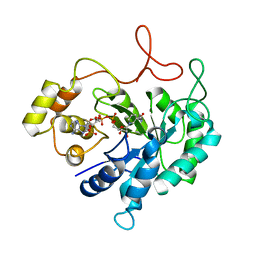 | | THE CRYSTAL STRUCTURE OF AN ALDEHYDE REDUCTASE Y50F MUTANT-NADP COMPLEX AND ITS IMPLICATIONS FOR SUBSTRATE BINDING | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Hyndman, D, Green, N.C, Li, L, Korithoski, B, Jia, Z, Flynn, T.G. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Aldehyde Reductase Y50F Mutant-NADP Complex and its Implications for Substrate Binding
Chem.Biol.Interact., 132, 2001
|
|
2R28
 
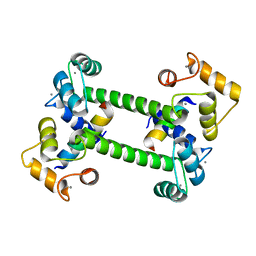 | |
2F2P
 
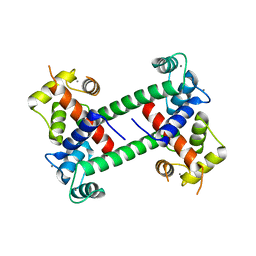 | |
6P8J
 
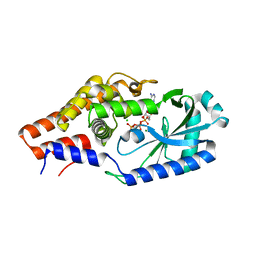 | |
6P8S
 
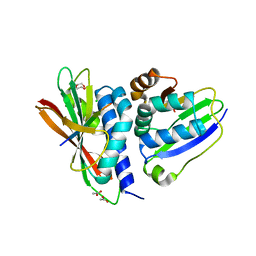 | |
6P8U
 
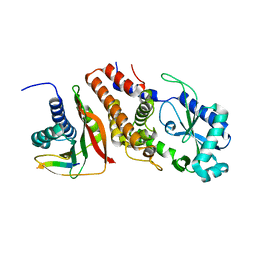 | | Structure of P. aeruginosa ATCC27853 CdnD:HORMA2:Peptide 1 complex | | Descriptor: | HORMA domain containing protein, MAGNESIUM ION, Nucleotidyltransferase, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
