5C9I
 
 | |
1J1W
 
 | | Crystal Structure Of The Monomeric Isocitrate Dehydrogenase In Complex With NADP+ | | Descriptor: | Isocitrate Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-12-19 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Monomeric Isocitrate Dehydrogenase in the Presence of NADP+
J.Biol.Chem., 278, 2003
|
|
1ITW
 
 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-02-12 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
4YFB
 
 | |
4YF9
 
 | |
4YFA
 
 | |
3B1N
 
 | | Structure of Burkholderia thailandensis nucleoside kinase (BthNK) in complex with ADP-mizoribine | | Descriptor: | 5-hydroxy-1-(beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yasutake, Y, Ota, H, Hino, E, Sakasegawa, S, Tamura, T. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Burkholderia thailandensis nucleoside kinase: implications for the catalytic mechanism and nucleoside selectivity
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3B1O
 
 | | Structure of Burkholderia thailandensis nucleoside kinase (BthNK) in ligand-free form | | Descriptor: | Ribokinase, putative | | Authors: | Yasutake, Y, Ota, H, Hino, E, Sakasegawa, S, Tamura, T. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Burkholderia thailandensis nucleoside kinase: implications for the catalytic mechanism and nucleoside selectivity
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5GNM
 
 | | Cytochrome P450 Vdh (CYP107BR1) L348M mutant | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Yasutake, Y, Tamura, T. | | Deposit date: | 2016-07-22 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5GNL
 
 | | Cytochrome P450 Vdh (CYP107BR1) F106V mutant | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Yasutake, Y, Tamura, T. | | Deposit date: | 2016-07-22 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XN2
 
 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
7COF
 
 | |
7COG
 
 | |
2DTX
 
 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with D-mannose | | Descriptor: | Glucose 1-dehydrogenase related protein, SULFATE ION, beta-D-mannopyranose | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-18 | | Release date: | 2007-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
2DTD
 
 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in ligand-free form | | Descriptor: | Glucose 1-dehydrogenase related protein, SULFATE ION | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-12 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
2DTE
 
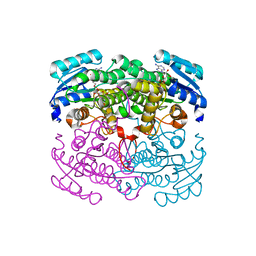 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase related protein | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-12 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
8X22
 
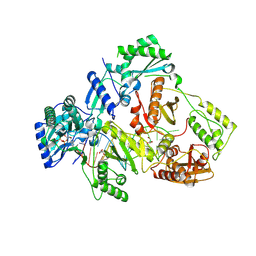 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8X21
 
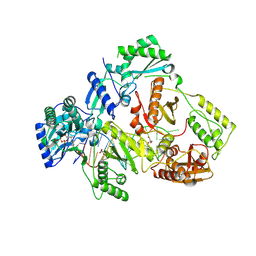 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:ETV-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8X1Z
 
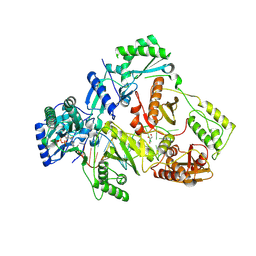 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8X20
 
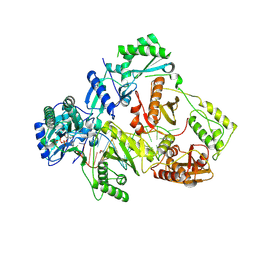 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
1WZZ
 
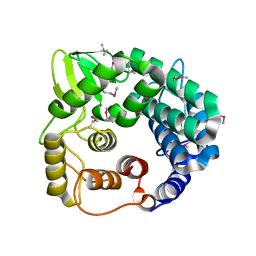 | | Structure of endo-beta-1,4-glucanase CMCax from Acetobacter xylinum | | Descriptor: | Probable endoglucanase, SULFATE ION | | Authors: | Yasutake, Y, Kawano, S, Tajima, K, Yao, M, Satoh, Y, Munekata, M, Tanaka, I, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the Acetobacter xylinum endo-beta-1,4-glucanase CMCax required for cellulose biosynthesis.
Proteins, 64, 2006
|
|
3VTZ
 
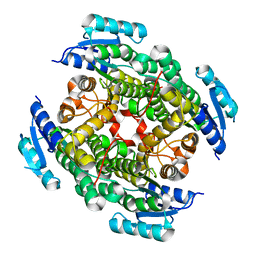 | |
3VUN
 
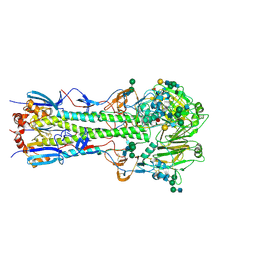 | | Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yasutake, Y, Suzuki, T, Kawaguchi, A, Nobusawa, E. | | Deposit date: | 2012-07-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group
To be Published
|
|
