1VGN
 
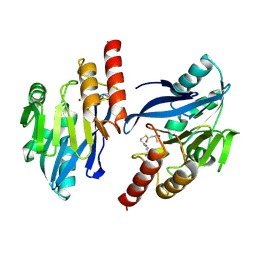 | | Structure-based design of the irreversible inhibitors to metallo--lactamase (IMP-1) | | Descriptor: | 3-OXO-3-[(3-OXOPROPYL)SULFANYL]PROPANE-1-THIOLATE, ACETIC ACID, Beta-lactamase IMP-1, ... | | Authors: | Kurosaki, H, Yamaguchi, Y, Higashi, T, Soga, K, Matsueda, S, Misumi, S, Yamagata, Y, Arakawa, Y, Goto, M. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Irreversible Inhibition of Metallo-beta-lactamase (IMP-1) by 3-(3-Mercaptopropionylsulfanyl)propionic Acid Pentafluorophenyl Ester
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
3AIH
 
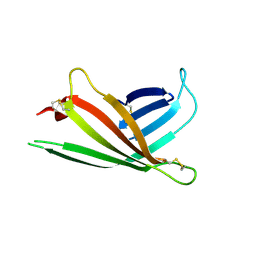 | | Human OS-9 MRH domain complexed with alpha3,alpha6-Man5 | | Descriptor: | Protein OS-9, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Satoh, T, Chen, Y, Hu, D, Hanashima, S, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2010-05-14 | | Release date: | 2010-12-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Oligosaccharide Recognition of Misfolded Glycoproteins by OS-9 in ER-Associated Degradation
Mol.Cell, 40, 2010
|
|
3APA
 
 | | Crystal structure of human pancreatic secretory protein ZG16p | | Descriptor: | CHLORIDE ION, GLYCEROL, Zymogen granule membrane protein 16 | | Authors: | Kanagawa, M, Satoh, T, Nakano, Y, Kojima-Aikawa, K, Yamaguchi, Y. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related beta-prism fold
Biochem.Biophys.Res.Commun., 2010
|
|
3AVE
 
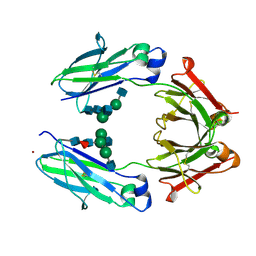 | | Crystal Structure of the Fucosylated Fc Fragment from Human Immunoglobulin G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, ZINC ION | | Authors: | Matsumiya, S, Yamaguchi, Y, Saito, J, Nagano, M, Sasakawa, H, Otaki, S, Satoh, M, Shitara, K, Kato, K. | | Deposit date: | 2011-03-04 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Corrigendum to "Structural Comparison of Fucosylated and Nonfucosylated Fc Fragments of Human Immunoglobulin G1" [J. Mol. Biol. 386/3 (2007) 767-779]
J.Mol.Biol., 408, 2011
|
|
3AQG
 
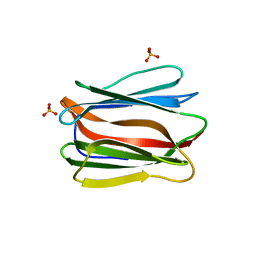 | | Crystal structure of human pancreatic secretory protein ZG16b | | Descriptor: | PHOSPHATE ION, Zymogen granule protein 16 homolog B | | Authors: | Kanagawa, M, Satoh, T, Nakano, Y, Kojima-Aikawa, K, Yamaguchi, Y. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related beta-prism fold
Biochem.Biophys.Res.Commun., 2010
|
|
3AQX
 
 | | Crystal structure of Bombyx mori beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, GLYCEROL, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQY
 
 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQZ
 
 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
5B1W
 
 | |
5AZW
 
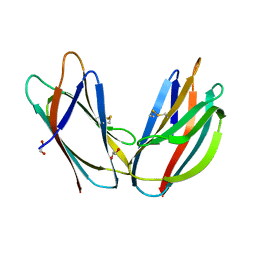 | | Crystal structure of p24beta1 GOLD domain | | Descriptor: | 1,2-ETHANEDIOL, Transmembrane emp24 domain-containing protein 2 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5AZY
 
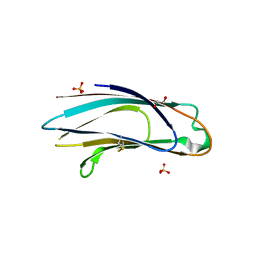 | | Crystal structure of p24delta1 GOLD domain (Native 2) | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5B1X
 
 | |
5AZX
 
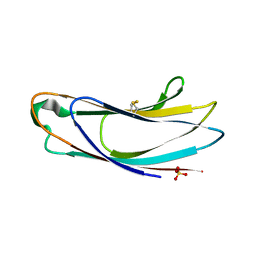 | | Crystal structure of p24delta1 GOLD domain (native 1) | | Descriptor: | SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UMH
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1WZ1
 
 | | Crystal structure of the Fv fragment complexed with dansyl-lysine | | Descriptor: | Ig heavy chain, Ig light chain, N~6~-{[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}-L-LYSINE | | Authors: | Nakasako, M, Oka, T, Mashumo, M, Takahashi, H, Shimada, I, Yamaguchi, Y, Kato, K, Arata, Y. | | Deposit date: | 2005-02-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational dynamics of complementarity-determining region H3 of an anti-dansyl Fv fragment in the presence of its hapten
J.Mol.Biol., 351, 2005
|
|
1WR6
 
 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1WRD
 
 | | Crystal structure of Tom1 GAT domain in complex with ubiquitin | | Descriptor: | Target of Myb protein 1, Ubiquitin | | Authors: | Akutsu, M, Kawasaki, M, Katoh, Y, Shiba, T, Yamaguchi, Y, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain.
Febs Lett., 579, 2005
|
|
2ZCB
 
 | | Crystal Structure of ubiquitin P37A/P38A | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Kitahara, R, Tanaka, T, Sakata, E, Yamaguchi, Y, Kato, K, Yokoyama, S. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of ubiquitin P37A/P38A
To be published
|
|
3A33
 
 | | UbcH5b~Ubiquitin Conjugate | | Descriptor: | GLYCEROL, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Sakata, E, Satoh, T, Yamamoto, S, Yamaguchi, Y, Yagi-Utsumi, M, Kurimoto, E, Wakatsuki, S, Kato, K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates
Structure, 18, 2010
|
|
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
5ZIC
 
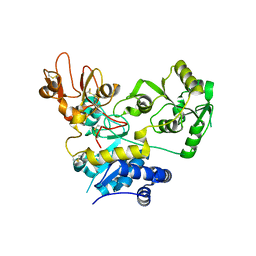 | | Crystal structure of human GnT-V luminal domain in complex with acceptor sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-6-thio-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V.
Nat Commun, 9, 2018
|
|
5ZIB
 
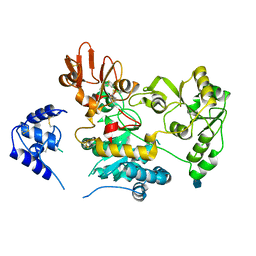 | | Crystal structure of human GnT-V luminal domain in apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V.
Nat Commun, 9, 2018
|
|
2D5U
 
 | |
2DPF
 
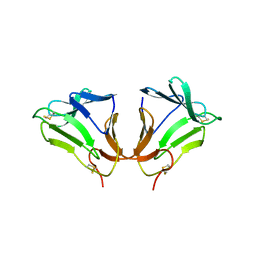 | | Crystal Structure of curculin1 homodimer | | Descriptor: | Curculin, SULFATE ION | | Authors: | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
