7YS8
 
 | |
7YST
 
 | |
7YS9
 
 | |
7YSY
 
 | |
7YSA
 
 | |
7YSM
 
 | |
7YT0
 
 | |
4RL4
 
 | |
7A73
 
 | |
8WZJ
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZM
 
 | |
8WZH
 
 | |
8WZK
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZI
 
 | | One RBD up state of Spike glycoprotein, SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8GR6
 
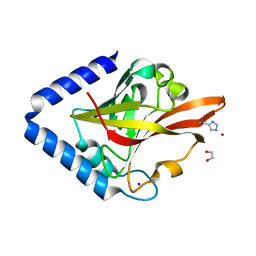 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
8GUU
 
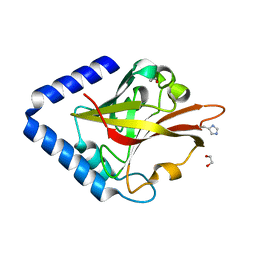 | |
8WV5
 
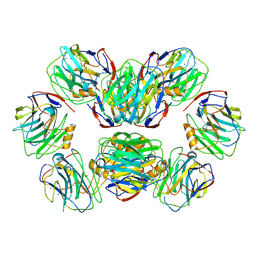 | | C-reactive protein, decamer | | Descriptor: | C-reactive protein(1-205), CALCIUM ION | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WV4
 
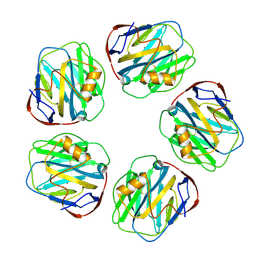 | | C-reactive protein, pentamer | | Descriptor: | C-reactive protein(1-205), CALCIUM ION | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WV6
 
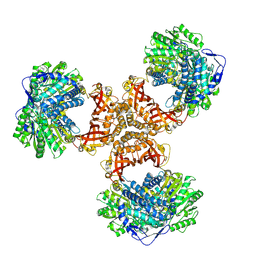 | | PaaZ, bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7W58
 
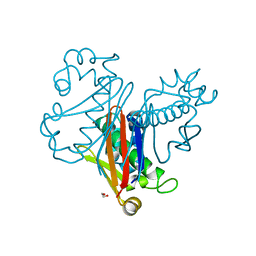 | | Crystal structure of acyl-carrier protein synthase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, 4'-phosphopantetheinyl transferase, NICKEL (II) ION | | Authors: | Yadav, S, Bhatia, I, Biswal, B.K. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification, structure determination and analysis of Mycobacterium smegmatis acyl-carrier protein synthase (AcpS) crystallized serendipitously.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
4GF9
 
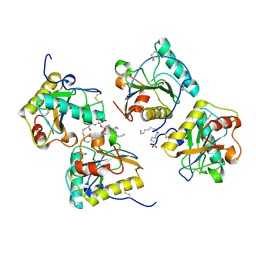 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
3ES6
 
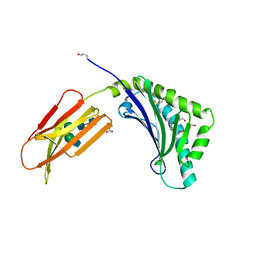 | | Crystal structure of the novel complex formed between Zinc 2-glycoprotein (ZAG) and Prolactin inducible protein (PIP) from human seminal plasma | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Hassan, M.I, Bilgrami, S, Kumar, V, Singh, N, Yadav, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal structure of the novel complex formed between zinc alpha2-glycoprotein (ZAG) and prolactin-inducible protein (PIP) from human seminal plasma
J.Mol.Biol., 384, 2008
|
|
6LVE
 
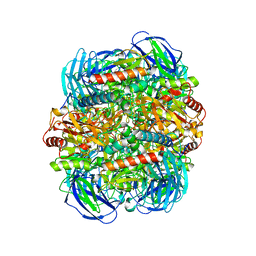 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
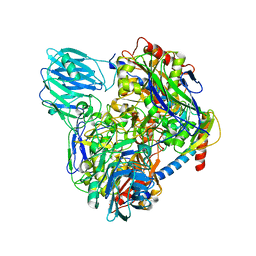 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVB
 
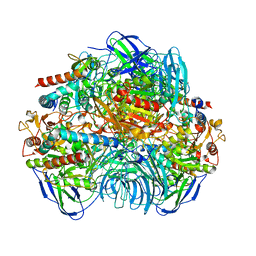 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
