5HTJ
 
 | |
5HV7
 
 | |
5HTX
 
 | |
7EPK
 
 | |
5ZYQ
 
 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
5ZYP
 
 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
7D99
 
 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D90
 
 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
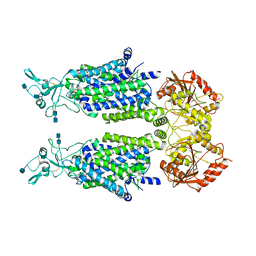 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
6E5U
 
 | |
3O4Z
 
 | |
4K6G
 
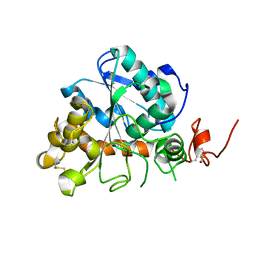 | | Crystal structure of CALB from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
6VHH
 
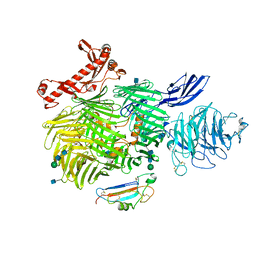 | | Human Teneurin-2 and human Latrophilin-3 binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L3, ... | | Authors: | Xie, Y, Li, J, Arac, D, Zhao, M. | | Deposit date: | 2020-01-09 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism.
Nat Commun, 11, 2020
|
|
2E0T
 
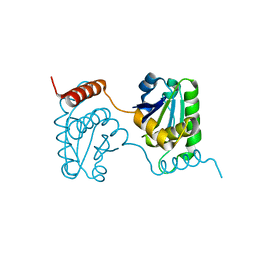 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | Descriptor: | Dual specificity phosphatase 26 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K6H
 
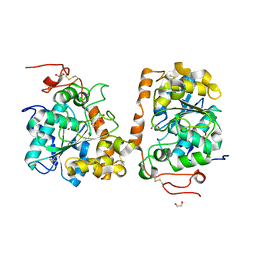 | | Crystal structure of CALB mutant L278M from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
4K5Q
 
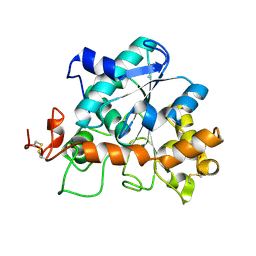 | |
4K6K
 
 | | Crystal structure of CALB mutant D223G from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-16 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
5KF4
 
 | |
6ABR
 
 | | Actin interacting protein 5 (Aip5, wild type) | | Descriptor: | Actin binding protein | | Authors: | Sun, J, Xie, Y, Toh, J.D.W, Hong, W, MIao, Y, Gao, Y.G. | | Deposit date: | 2018-07-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
6M4C
 
 | | C. albicans actin interacting protein Aip5 | | Descriptor: | C. albicans actin interacting protein Aip5 | | Authors: | Loh, Z.Y, Gao, Y.G, Xie, Y, Miao, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Orchestrated actin nucleation by the Candida albicans polarisome complex enables filamentous growth.
J.Biol.Chem., 295, 2020
|
|
1F03
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1IWA
 
 | | RUBISCO FROM GALDIERIA PARTITA | | Descriptor: | SULFATE ION, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | | Authors: | Okano, Y, Mizohata, E, Xie, Y, Matsumura, H, Sugawara, H, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2002-04-30 | | Release date: | 2003-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Galdieria Rubisco Complexed with one sulfate ion per active site
FEBS LETT., 527, 2002
|
|
1ES1
 
 | | CRYSTAL STRUCTURE OF VAL61HIS MUTANT OF TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
1EHB
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | PROTEIN (CYTOCHROME B5), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-02-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
