6JFH
 
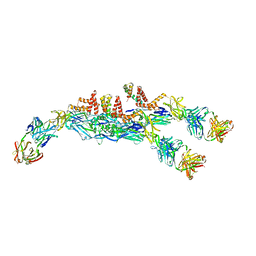 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
7K3A
 
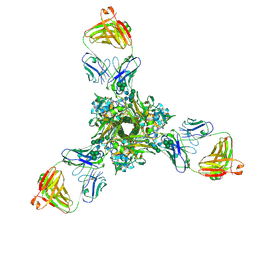 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation B, fusion peptide release | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3B
 
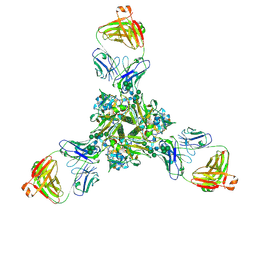 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation C, central helices splay | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K37
 
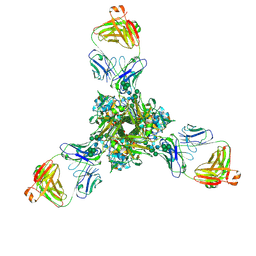 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K39
 
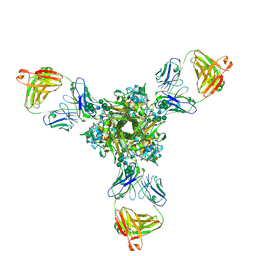 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7FFO
 
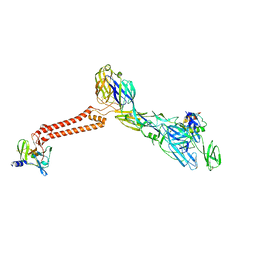 | | Cryo-EM structure of VEEV VLP at the 5-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFQ
 
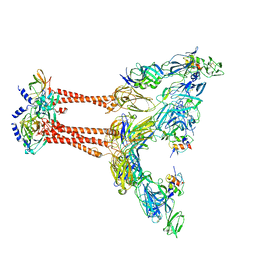 | | Cryo-EM structure of VEEV VLP at the 2-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFL
 
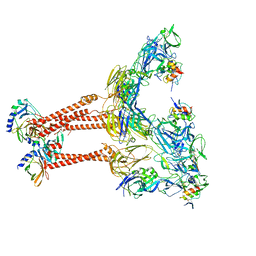 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 2-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFE
 
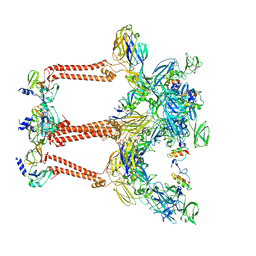 | | Cryo-EM structure of VEEV VLP | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFF
 
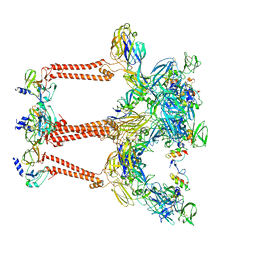 | | Structure of Venezuelan equine encephalitis virus with the receptor LDLRAD3 | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFN
 
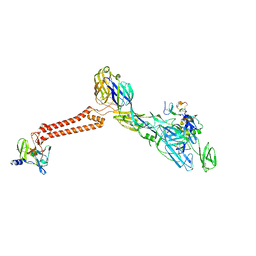 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 5-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
6A7F
 
 | |
8IGL
 
 | |
8IGG
 
 | |
7XY1
 
 | |
7XYC
 
 | |
7Y3T
 
 | |
7Y22
 
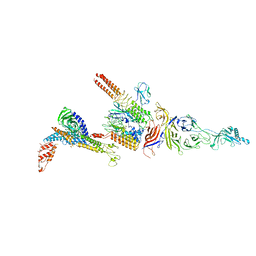 | |
7Y23
 
 | |
7Y1C
 
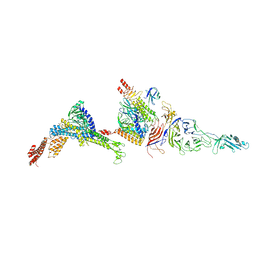 | |
7Y5S
 
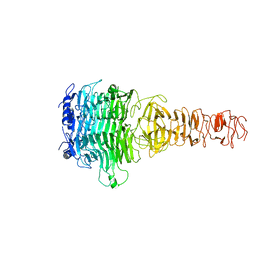 | |
8IHP
 
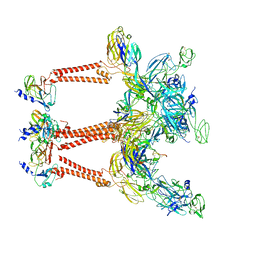 | | Structure of Semliki Forest virus VLP in complex with the receptor VLDLR-LA3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Zhang, X, Xiang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Semliki Forest virus in complex with its receptor VLDLR.
Cell, 186, 2023
|
|
7V7C
 
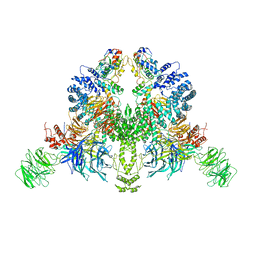 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | Authors: | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
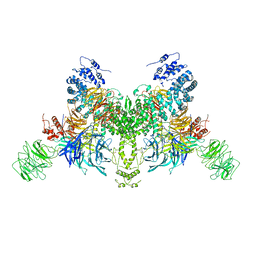
7V7B
 
 | |
7W68
 
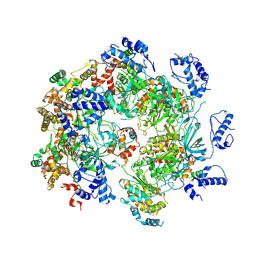 | | human single hexameric Mcm2-7 complex | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Xu, N.N, Lin, Q.P, Liu, C.D, Tian, H.L, Xiang, Y, Zhu, G. | | Deposit date: | 2021-12-01 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | human single hexameric Mcm2-7 complex
To Be Published
|
|
