5T4L
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | 分子名称: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | 著者 | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | 登録日 | 2016-08-29 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4K
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | 分子名称: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | 著者 | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | 登録日 | 2016-08-29 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4N01
 
 | |
4RBN
 
 | | The crystal structure of Nitrosomonas europaea sucrose synthase: Insights into the evolutionary origin of sucrose metabolism in prokaryotes | | 分子名称: | Sucrose synthase:Glycosyl transferases group 1 | | 著者 | Wu, R, Asencion Diez, M.D, Figueroa, C.M, Machtey, M, Iglesias, A.A, Ballicora, M.A, Liu, D. | | 登録日 | 2014-09-12 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
J.Bacteriol., 197, 2015
|
|
4RJ0
 
 | |
4RIZ
 
 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-07 | | 公開日 | 2014-10-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
4RM7
 
 | |
4RT5
 
 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from planctomyces limnophilus dsm 3776 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | 著者 | Wu, R, Bearden, J, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-13 | | 公開日 | 2014-12-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from Planctomyces limnophilus dsm 3776
TO BE PUBLISHED
|
|
4RU0
 
 | | The crystal structure of abc transporter permease from pseudomonas fluorescens group | | 分子名称: | 3,6,9,12,15-PENTAOXAHEPTADECANE, GLYCEROL, Putative branched-chain amino acid ABC transporter, ... | | 著者 | Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-17 | | 公開日 | 2014-11-26 | | 実験手法 | X-RAY DIFFRACTION (2.442 Å) | | 主引用文献 | The crystal structure of abc transporter permease from pseudomonas fluorescens group
To be Published
|
|
4RM1
 
 | |
4RLG
 
 | |
4M88
 
 | |
4MAA
 
 | |
4Y0I
 
 | | GABA-aminotransferase inactivated by conformationally-restricted inactivator | | 分子名称: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, 4-aminobutyrate aminotransferase, mitochondrial, ... | | 著者 | Wu, R, Sanishvili, R, Lee, H.V, Dustin, D.H, Emma, D, Neil, K, Silverman, R.B, Liu, D. | | 登録日 | 2015-02-06 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.662 Å) | | 主引用文献 | GABA-aminotransferase inactivated by conformationally-restricted inactivator
To Be Published
|
|
1RZ2
 
 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | 分子名称: | conserved hypothetical protein BA4783 | | 著者 | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-12-23 | | 公開日 | 2004-07-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
1SQE
 
 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | 分子名称: | hypothetical protein PG130 | | 著者 | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-03-18 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
4WKS
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | Acyl-homoserine lactone acylase PvdQ, ethylboronic acid | | 著者 | Wu, R, Clevenger, D.K, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.629 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
4WKV
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, trihydroxy(octyl)borate(1-) | | 著者 | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1434 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
4WKU
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, hexyl(trihydroxy)borate(1-) | | 著者 | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
4WKT
 
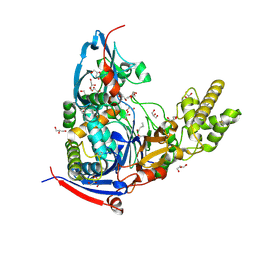 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | 1-BUTANE BORONIC ACID, Acyl-homoserine lactone acylase PvdQ, GLYCEROL | | 著者 | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.782 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
2OQW
 
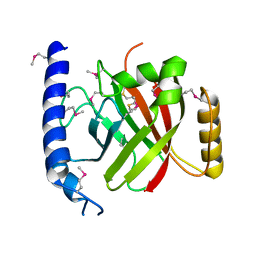 | | The crystal structure of sortase B from B.anthracis in complex with AAEK1 | | 分子名称: | Sortase B | | 著者 | Wu, R, Zhang, R, Marresso, A.W, Schneewind, O, Joachimiak, A. | | 登録日 | 2007-02-01 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2OQZ
 
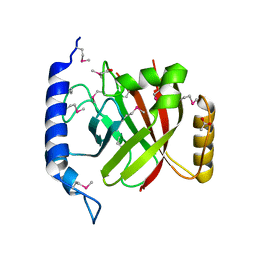 | | The crystal structure of sortase B from B.anthracis in complex with AAEK2 | | 分子名称: | ACETIC ACID, Sortase B | | 著者 | Wu, R, Zhang, R, Maresso, A.W, Schneewind, O, Joachimiak, A. | | 登録日 | 2007-02-01 | | 公開日 | 2007-06-19 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
1XBW
 
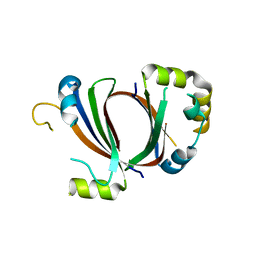 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | 分子名称: | hypothetical protein isdG | | 著者 | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-08-31 | | 公開日 | 2004-10-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
7TVL
 
 | | Viral AMG chitosanase V-Csn, apo structure | | 分子名称: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | 著者 | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | 登録日 | 2022-02-05 | | 公開日 | 2022-10-05 | | 実験手法 | X-RAY DIFFRACTION (0.89 Å) | | 主引用文献 | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
