3LDU
 
 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3LAX
 
 | |
3QSJ
 
 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3QSG
 
 | |
3RQZ
 
 | | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Metallophosphoesterase, ZINC ION | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus
To be Published
|
|
2FUV
 
 | | Phosphoglucomutase from Salmonella typhimurium. | | Descriptor: | MAGNESIUM ION, phosphoglucomutase | | Authors: | Osipiuk, J, Wu, R, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of phosphoglucomutase from Salmonella typhimurium.
To be Published
|
|
2GBO
 
 | |
2GRE
 
 | |
2IDL
 
 | | Crystal Structure of Conserved Protein of Unknown Function from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, Hypothetical protein, SODIUM ION | | Authors: | Nocek, B, Wu, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
4ESY
 
 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
4GAK
 
 | | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale | | Descriptor: | Acyl-ACP thioesterase, CHLORIDE ION, GLYCEROL | | Authors: | Chang, C, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale
To be Published
|
|
5TTX
 
 | |
2NN5
 
 | | Structure of Conserved Protein of Unknown Function EF2215 from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, Hypothetical protein EF_2215, MAGNESIUM ION | | Authors: | Osipiuk, J, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein EF_2215 from Enterococcus faecalis.
To be Published
|
|
2OCD
 
 | | Crystal structure of L-asparaginase I from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, L-asparaginase I | | Authors: | Nocek, B, Wu, R, Osipiuk, J, Moy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-asparaginase I from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
2OEQ
 
 | |
2OQT
 
 | | Structural Genomics, the crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS | | Descriptor: | Hypothetical protein SPy0176 | | Authors: | Tan, K, Wu, R, Osipiuk, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS
To be Published
|
|
4I6V
 
 | | The crystal structure of an amidohydrolase 2 from Planctomyces limnophilus DSM 3776 | | Descriptor: | ACETATE ION, Amidohydrolase 2, GLYCEROL, ... | | Authors: | Fan, Y, Tan, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | The crystal structure of an amidohydrolase 2 from Planctomyces limnophilus DSM 3776
To be Published
|
|
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-07-17 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
2QYT
 
 | | Crystal structure of 2-dehydropantoate 2-reductase from Porphyromonas gingivalis W83 | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydropantoate 2-reductase, SULFATE ION | | Authors: | Tan, K, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-15 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of 2-dehydropantoate 2-reductase from Porphyromonas gingivalis W83.
To be Published
|
|
2QQY
 
 | | Crystal structure of ferritin like, diiron-carboxylate proteins from Bacillus anthracis str. Ames | | Descriptor: | Sigma B operon | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Ferritin like, Diiron-carboxylate Proteins from Bacillus anthracis str. Ames.
To be Published
|
|
2R2A
 
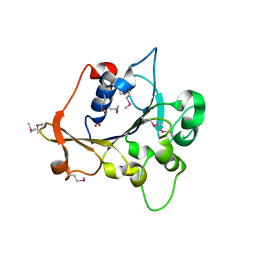 | | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Patterson, S, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-24 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis.
To be Published
|
|
2QQZ
 
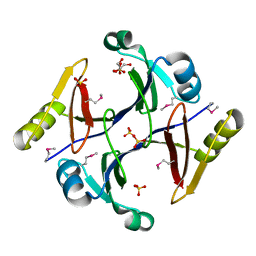 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
2R5R
 
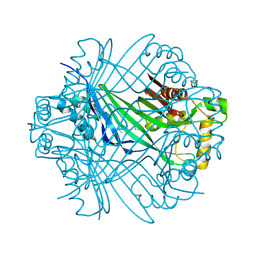 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
2R5S
 
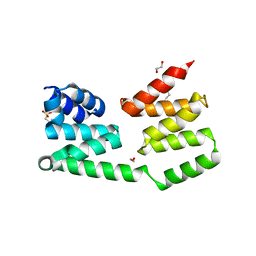 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
4KTB
 
 | | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Tan, K, Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603
To be Published
|
|
