2ONY
 
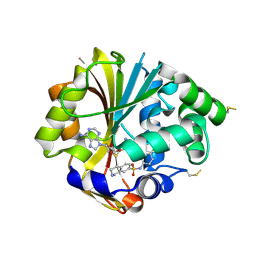 | | Structure of hPNMT with inhibitor 7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | N-(4-CHLOROPHENYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-24 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
7DL3
 
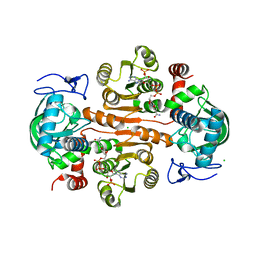 | | The structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84606934 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL0
 
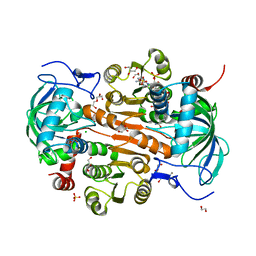 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
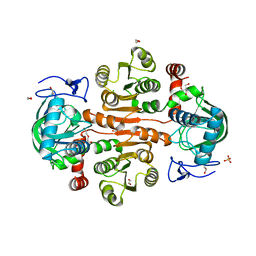 | | The wild-type structure of 3,5-DAHDHcca | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.30065823 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL1
 
 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6M53
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
7EQL
 
 | | Crystal structure of (+)-pulegone reductase from Mentha piperita | | Descriptor: | (+)-pulegone reductase | | Authors: | Lin, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Functional Characterization and Structural Insights Into Stereoselectivity of Pulegone Reductase in Menthol Biosynthesis.
Front Plant Sci, 12, 2021
|
|
6IFE
 
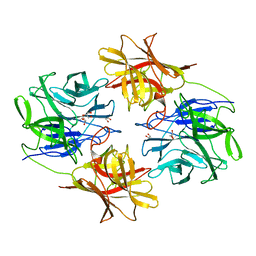 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
4HRI
 
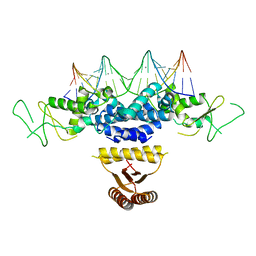 | | Crystal structure of HetR in complex with a 21-bp palindromic DNA at the upstream of the hetP promoter from Anabaena | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Chen, Y, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2012-10-28 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural and biochemical analyses of Anabaena HetR reveal insights into its binding to DNA targets and the inhibitory hexapeptide ERGSGR
To be Published
|
|
1N9U
 
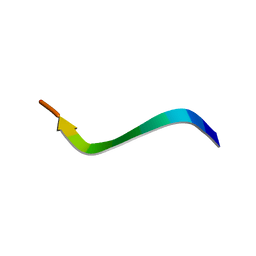 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship | | Descriptor: | Angiotensin I | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
4LH9
 
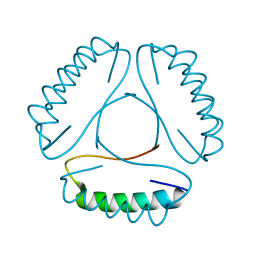 | |
1N9V
 
 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship. | | Descriptor: | Angiotensin II | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
4NBM
 
 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NAA
 
 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | Deposit date: | 2013-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NC4
 
 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
5YP6
 
 | | RORgamma (263-509) complexed with SRC2 and Compound 6 | | Descriptor: | N-[3'-cyano-4'-(2-methylpropyl)-2-(trifluoromethyl)biphenyl-4-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, SRC2 | | Authors: | Gao, M, Cai, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
5YP5
 
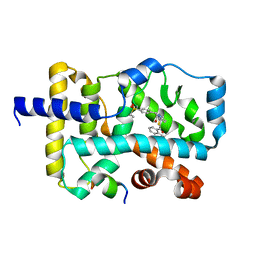 | | Crystal structure of RORgamma complexed with SRC2 and compound 5d | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-{5-[2-(2-methylpropyl)benzoyl]-4-phenyl-1,3-thiazol-2-yl}acetamide, Nuclear receptor ROR-gamma, SRC2-2 peptide | | Authors: | Gao, M, Cai, W, Chunwa, C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
5UHT
 
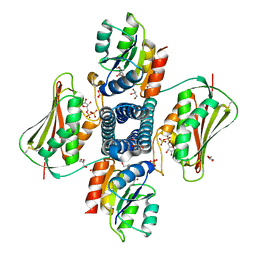 | | Structure of the Thermotoga maritima HK853-BeF3-RR468 complex at pH 5.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, Y, Rose, J, Jiang, L, Zhou, P. | | Deposit date: | 2017-01-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
6NM3
 
 | | NMR structure of WW295 | | Descriptor: | WW295 peptide | | Authors: | Wang, G, Zarena, D. | | Deposit date: | 2019-01-10 | | Release date: | 2020-07-15 | | Last modified: | 2020-09-09 | | Method: | SOLUTION NMR | | Cite: | Two distinct amphipathic peptide antibiotics with systemic efficacy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NM4
 
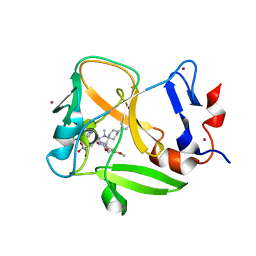 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | Descriptor: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | Authors: | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
