4DNW
 
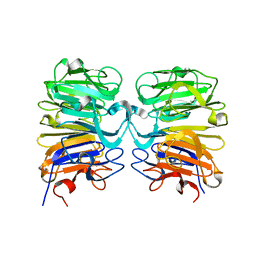 | | Crystal structure of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
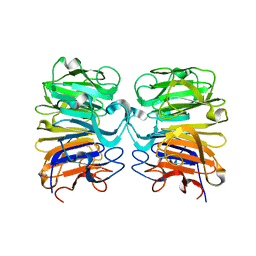 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
7ZDC
 
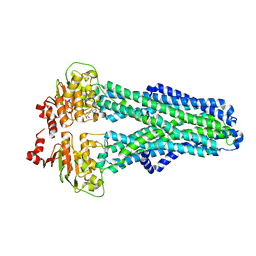 | |
7ZDA
 
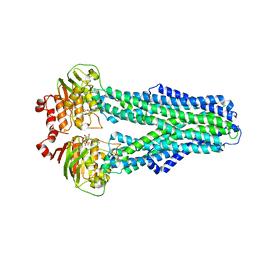 | |
7ZDK
 
 | |
7ZDB
 
 | |
7ZDE
 
 | |
7ZDS
 
 | |
7ZDV
 
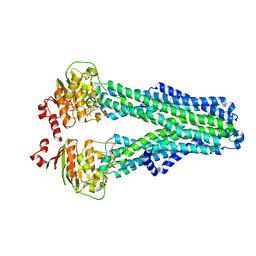 | |
7ZDW
 
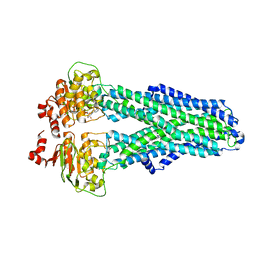 | |
7ZEC
 
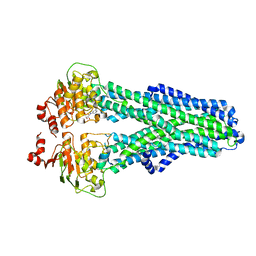 | |
7ZDG
 
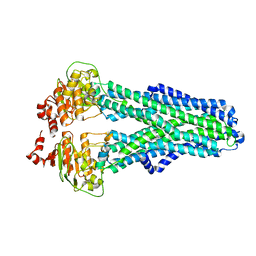 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | 分子名称: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | 著者 | Wu, D, Safarian, S. | | 登録日 | 2022-03-29 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZD5
 
 | |
7ZDL
 
 | |
7ZDT
 
 | |
7ZDR
 
 | |
7ZE5
 
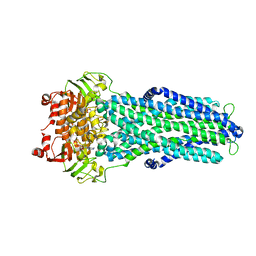 | |
7ZDF
 
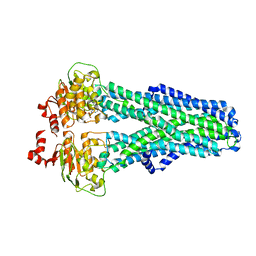 | |
7ZDU
 
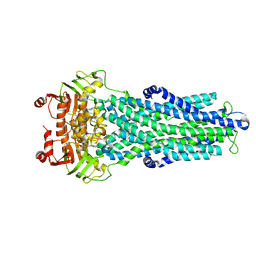 | |
5WTI
 
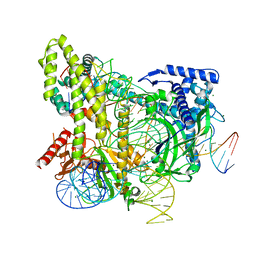 | | Crystal structure of the CRISPR-associated protein in complex with crRNA and DNA | | 分子名称: | CRISPR-associated protein, DNA (28-MER), DNA (5'-D(P*GP*TP*GP*TP*GP*GP*AP*TP*TP*CP*CP*G)-3'), ... | | 著者 | Wu, D, Guan, X, Zhu, Y, Huang, Z. | | 登録日 | 2016-12-13 | | 公開日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.682 Å) | | 主引用文献 | Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA
Cell Res., 27, 2017
|
|
5XFJ
 
 | |
5XFF
 
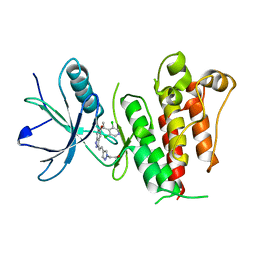 | |
5ZZ2
 
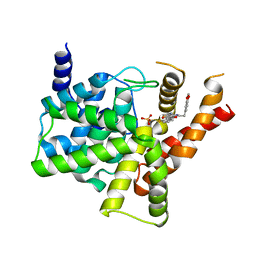 | | Crystal structure of PDE5 in complex with inhibitor LW1634 | | 分子名称: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2018-05-29 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
6ACB
 
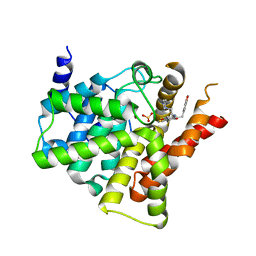 | | Crystal structure of PDE5 in complex with inhibitor LW1805 | | 分子名称: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-6-{[2-(4-methylpiperazin-1-yl)ethyl]amino}-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2018-07-26 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
