5FRV
 
 | |
5FRZ
 
 | |
2WCS
 
 | | Crystal Structure of Debranching enzyme from Nostoc punctiforme (NPDE) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.B, Choi, J.H, Nam, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
2WKG
 
 | | Nostoc punctiforme Debranching Enzyme (NPDE)(Native form) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.B, Choi, J.H, Song, H.N, Park, K.H, Woo, E.J. | | Deposit date: | 2009-06-11 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
4UW2
 
 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
7VT9
 
 | |
3K1I
 
 | | Crystal strcture of FliS-HP1076 complex in H. pylori | | Descriptor: | Flagellar protein, Putative uncharacterized protein | | Authors: | Lam, W.W.L, Kotaka, M, Ling, T.K.W, Au, S.W.N. | | Deposit date: | 2009-09-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular interaction of flagellar export chaperone FliS and cochaperone HP1076 in Helicobacter pylori
Faseb J., 24, 2010
|
|
2E0T
 
 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | Descriptor: | Dual specificity phosphatase 26 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5G4D
 
 | |
5YSZ
 
 | | transcriptional regulator CelR-cellobiose complex | | Descriptor: | GLYCEROL, Transcriptional regulator, LacI family, ... | | Authors: | Fu, Y, Yeom, S.Y, Lee, D.H, Lee, S.G. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structural and functional analyses of the cellulase transcription regulator CelR
FEBS Lett., 592, 2018
|
|
3EZZ
 
 | | Crystal Structure of human MKP-2 | | Descriptor: | Dual specificity protein phosphatase 4, SULFATE ION | | Authors: | Jeong, D.G, Jung, S.K, Ryu, S.E, Kim, S.J. | | Deposit date: | 2008-10-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the catalytic domain of human MKP-2 reveals a 24-mer assembly.
Proteins, 76, 2009
|
|
5FS0
 
 | |
8IIS
 
 | | MsmUdgX H109S/R184A double mutant | | Descriptor: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIO
 
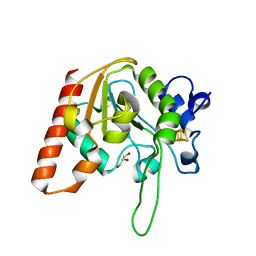 | | H109Q mutant of uracil DNA glycosylase X | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIN
 
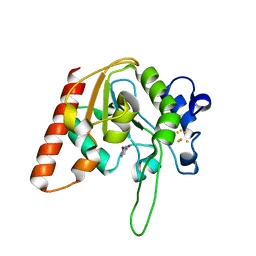 | |
8IIM
 
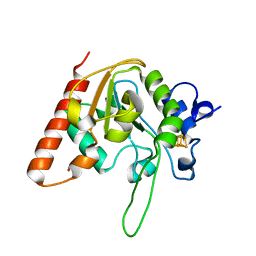 | | H109K mutant of uracil DNA glycosylase X | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIP
 
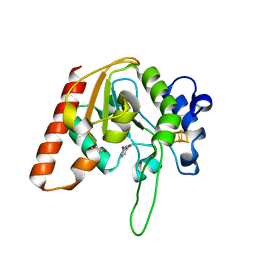 | |
8IIF
 
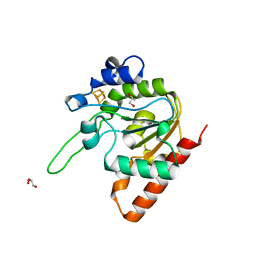 | | H109A mutant of uracil DNA glycosylase X | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIT
 
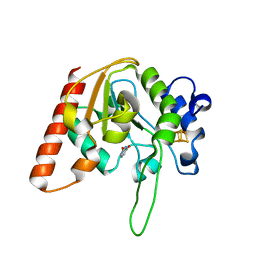 | |
8IIL
 
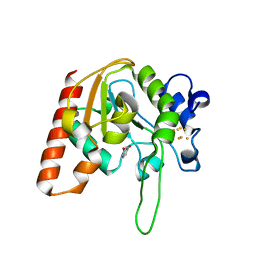 | |
8IIJ
 
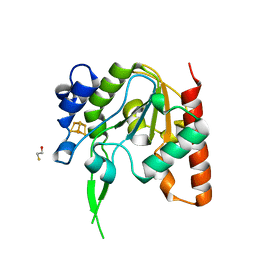 | | H109G mutant of uracil DNA glycosylase X | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIE
 
 | |
8IIQ
 
 | | MsmUdgX H109S/E52N double mutant | | Descriptor: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIG
 
 | |
8IIR
 
 | | MsmUdgX H109S/Q53A double mutant | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
