7KWW
 
 | | X-ray Crystal Structure of PlyCB Mutant K59H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWT
 
 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | Descriptor: | PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7LZ6
 
 | | The Cryo-EM structure of a complex between GAD65 and b96.11 Fab | | Descriptor: | Glutamate decarboxylase 2, b96.11 Fab heavy chain, b96.11 Fab light chain | | Authors: | Reboul, C.F, Le, S.N, Williams, D.E, Buckle, A.M. | | Deposit date: | 2021-03-09 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and dynamics of the autoantigen GAD65 in complex with the human autoimmune polyendocrine syndrome type 2-associated autoantibody b96.11
Biorxiv, 2024
|
|
2JUY
 
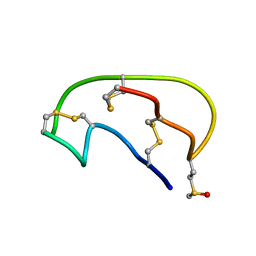 | | NMR ensemble of Neopetrosiamide A | | Descriptor: | Neopetrosiamide A | | Authors: | Austin, P, Williams, D.E, Heller, M, McIntosh, L.P, Andersen, R.J, Roberge, M, Roskelley, C.D. | | Deposit date: | 2007-09-05 | | Release date: | 2008-08-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR ensemble of Neopetrosiamide A
To be Published
|
|
4W93
 
 | | Human pancreatic alpha-amylase in complex with montbretin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Montbretin A, ... | | Authors: | Williams, L.K, Caner, S, Brayer, G.D. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Nat.Chem.Biol., 11, 2015
|
|
4YV8
 
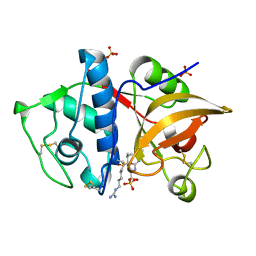 | | Crystal structure of cathepsin K bound to the covalent inhibitor lichostatinal | | Descriptor: | Cathepsin K, Lichostatinal, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
4YVA
 
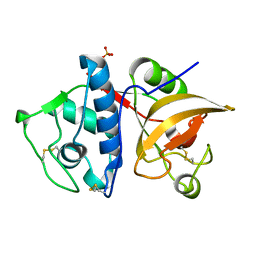 | | Cathepsin K co-crystallized with actinomycetes extract | | Descriptor: | Cathepsin K, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
