7B3K
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
4WZG
 
 | | Structure of human ATG101 | | Descriptor: | Autophagy-related protein 101, BETA-MERCAPTOETHANOL | | Authors: | Michel, M, Weiergraeber, O.H. | | Deposit date: | 2014-11-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mammalian autophagy initiator complex contains 2 HORMA domain proteins.
Autophagy, 11, 2015
|
|
4XTL
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric blue form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, SODIUM ION, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XOK
 
 | | Observing the overall rocking motion of a protein in a crystal. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Ma, P, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
4XOL
 
 | | Observing the overall rocking motion of a protein in a crystal - Cubic Ubiquitin crystals. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Peixiang, M, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
4XOF
 
 | |
4XTN
 
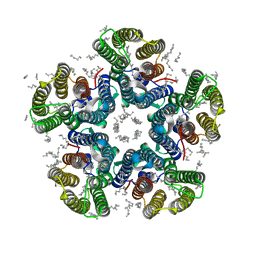 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 4.9 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTO
 
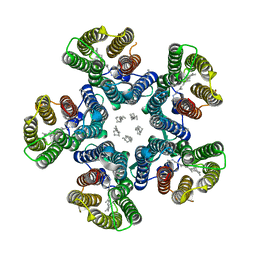 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 5.6 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AHT
 
 | |
6FXF
 
 | |
6G8O
 
 | |
7ONY
 
 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONO
 
 | |
7ONW
 
 | | Crystal structure of PBP3 from E. coli in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONK
 
 | | Crystal structure of PBP3 from P. aeruginosa in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONN
 
 | | Crystal structure of PBP3 transpeptidase domain from E. coli in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, DODECAETHYLENE GLYCOL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONX
 
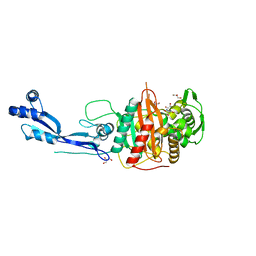 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONZ
 
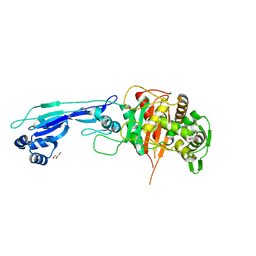 | |
4MD2
 
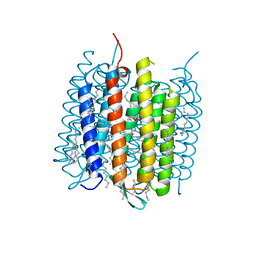 | | Ground state of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6EID
 
 | | Crystal structure of wild-type Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, PHOSPHATE ION, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
6EIG
 
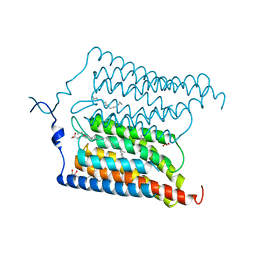 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | Authors: | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
4MD1
 
 | | Orange species of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7YX0
 
 | | Crystal structure of the full-length short LOV protein SBW25-LOV from Pseudomonas fluorescens (light state) | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Flavin mononucleotide (semi-quinone intermediate), ... | | Authors: | Arinkin, V, Batra-Safferling, R, Granzin, J. | | Deposit date: | 2022-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved Signal Transduction Mechanisms and Dark Recovery Kinetic Tuning in the Pseudomonadaceae Short Light, Oxygen, Voltage (LOV) Protein Family.
J.Mol.Biol., 2024
|
|
6G8C
 
 | |
6G8E
 
 | |
