1TX2
 
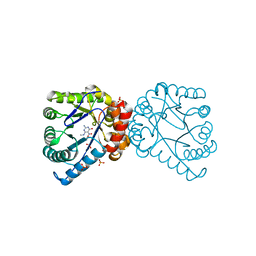 | | Dihydropteroate Synthetase, With Bound Inhibitor MANIC, From Bacillus anthracis | | Descriptor: | 6-METHYLAMINO-5-NITROISOCYTOSINE, DHPS, Dihydropteroate synthase, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.E, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
4M5L
 
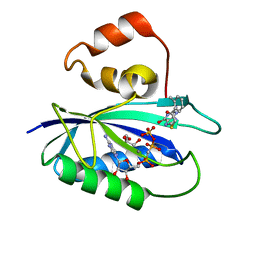 | | The Identification, Analysis and Structure-Based Development of Novel Inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-amino-8-{[2-(2-methylphenyl)-2-oxoethyl]sulfanyl}-1,7-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Yun, M, Hoagland, D, Kumar, G, Waddell, B, Rock, C.O, Lee, R.E, White, S.W. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.094 Å) | | Cite: | The identification, analysis and structure-based development of novel inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.
Bioorg.Med.Chem., 22, 2014
|
|
4M5K
 
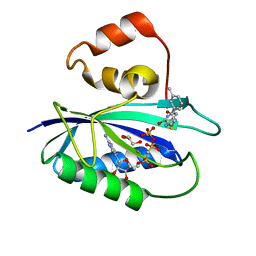 | | The Identification, Analysis and Structure-Based Development of Novel Inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-amino-8-{[2-(3-methylphenyl)-2-oxoethyl]sulfanyl}-1,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Yun, M, Hoagland, D, Kumar, G, Waddell, B, Rock, C.O, Lee, R.E, White, S.W. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | The identification, analysis and structure-based development of novel inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.
Bioorg.Med.Chem., 22, 2014
|
|
4N8M
 
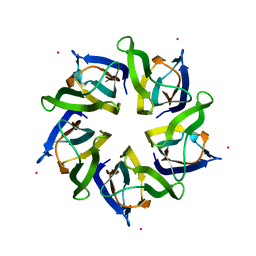 | | Structural polymorphism in the N-terminal oligomerization domain of NPM1 | | Descriptor: | COBALT (II) ION, Nucleophosmin | | Authors: | Mitrea, D, Royappa, G, Buljan, M, Yun, M, Pytel, N, Satumba, J, Nourse, A, Park, C, Babu, M.M, White, S.W, Kriwacki, R.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural polymorphism in the N-terminal oligomerization domain of NPM1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NHV
 
 | |
4NIL
 
 | |
4NIR
 
 | |
4NL1
 
 | |
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
1QDN
 
 | | AMINO TERMINAL DOMAIN OF THE N-ETHYLMALEIMIDE SENSITIVE FUSION PROTEIN (NSF) | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (N-ETHYLMALEIMIDE SENSITIVE FUSION PROTEIN (NSF)), SULFATE ION | | Authors: | May, A.P, Misura, K.M.S, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1999-05-21 | | Release date: | 1999-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein.
Nat.Cell Biol., 1, 1999
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
1BM9
 
 | |
4X9X
 
 | | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid Binding Protein Family | | Descriptor: | 1,2-ETHANEDIOL, DegV domain-containing protein MW1315, OLEIC ACID | | Authors: | Broussard, T.C, Miller, D.J, Jackson, P, Nourse, A, Rock, C.O. | | Deposit date: | 2014-12-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid-binding Protein Family.
J.Biol.Chem., 291, 2016
|
|
487D
 
 | |
5W9G
 
 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9k (SRI-30023) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4 -carboxamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W73
 
 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9f (SRI-29835) | | Descriptor: | 2-{(2S)-1-[(2-chlorophenoxy)acetyl]pyrrolidin-2-yl}-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W92
 
 | | Crystal structure of the influenza virus PA endonuclease in complex with an inhibitor - SRI-30049 | | Descriptor: | 1-[(3R,5S,7R)-1,5,7,9-tetrakis(2-oxopyrrolidin-1-yl)nonan-3-yl]-1,3-dihydro-2H-pyrrol-2-one, 2-[(2S)-1-(3,5-dichloropyridine-4-carbonyl)pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-[2-(phenylsulfonyl)ethyl]-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W7U
 
 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 8f (SRI-29928) | | Descriptor: | 2-[(2S)-1-(3,5-dichloropyridine-4-carbonyl)pyrrolidin-2-yl]-N-(2,3-dihydro-1H-inden-2-yl)-5-hydroxy-6-oxo-1,6-dihydropy rimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
6NXD
 
 | |
3IO5
 
 | |
2M03
 
 | |
2M04
 
 | |
6NX9
 
 | |
6NXB
 
 | | ECAII IN COMPLEX WITH CITRATE AT PH 7 | | Descriptor: | CITRIC ACID, GLYCEROL, L-asparaginase 2 | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2019-02-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Sci Rep, 9, 2019
|
|
