1URF
 
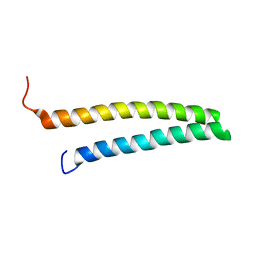 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
2QNA
 
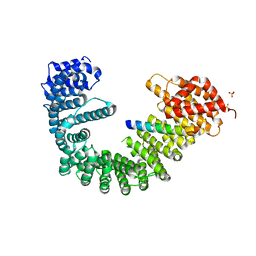 | | Crystal structure of human Importin-beta (127-876) in complex with the IBB-domain of Snurportin1 (1-65) | | Descriptor: | Importin subunit beta-1, SULFATE ION, Snurportin-1 | | Authors: | Wohlwend, D, Strasser, A, Dickmanns, A, Ficner, R. | | Deposit date: | 2007-07-18 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus.
J.Mol.Biol., 374, 2007
|
|
4LYS
 
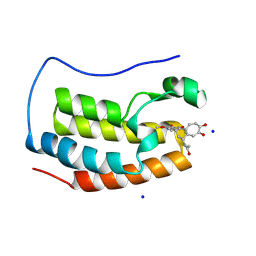 | | Crystal Structure of BRD4(1) bound to Colchiceine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-10-hydroxy-1,2,3-trimethoxy-9-oxo-5,6,7,9-tetrahydrobenzo[a]heptalen-7-yl]acetamide, SODIUM ION | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6Q9C
 
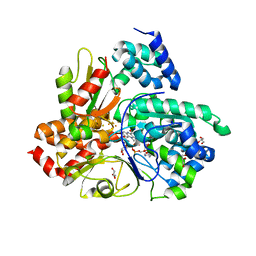 | | Crystal structure of Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF bound to NADH under anaerobic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9G
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129D and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9J
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129S and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9K
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF S96M bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
9FII
 
 | | Crystal Structure of oxidized NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE7
 
 | | Crystal Structure of oxidized NuoEF variant P228R(NuoF) from Aquifex aeolicus | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIH
 
 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions after 10 min soaking | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FDK
 
 | | Crystal Structure of oxidized NuoEF variant R66G(NuoF) from Aquifex aeolicus | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FEA
 
 | | Crystal Structure of reduced NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE0
 
 | | Crystal Structure of reduced NuoEF variant R66G(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FDJ
 
 | | Crystal structure of the NuoEF variant R66G (NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions (short soak) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FDV
 
 | | Crystal Structure of reduced NuoEF variant R66G(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIJ
 
 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE5
 
 | | Crystal Structure of NuoEF variant R66G(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions after 10 min soaking | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE8
 
 | | Crystal Structure of reduced NuoEF variant P228R(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIL
 
 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIF
 
 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
5D3J
 
 | | First bromodomain of BRD4 bound to inhibitor XD33 | | Descriptor: | 4-acetyl-N-[3-(diethylsulfamoyl)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3R
 
 | | First bromodomain of BRD4 bound to inhibitor XD42 | | Descriptor: | 4-acetyl-N-[3-(azepan-1-ylsulfonyl)phenyl]-5-methyl-3-propyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D26
 
 | | First bromodomain of BRD4 bound to inhibitor XD28 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D25
 
 | | First bromodomain of BRD4 bound to inhibitor XD27 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D24
 
 | | First bromodomain of BRD4 bound to inhibitor XD26 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-[3-(2-amino-2-oxoethoxy)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
