7B81
 
 | |
7CGR
 
 | |
7CGQ
 
 | |
7DO6
 
 | |
7DO5
 
 | |
7DO7
 
 | |
7YAX
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, | | Descriptor: | CHLORIDE ION, Hydroxynitrile lyase, SULFATE ION | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based site-directed mutagenesis of hydroxynitrile lyase from cyanogenic millipede, Oxidus gracilis for enhancing catalytic efficiency and enantioselectivity
To Be Published
|
|
7YCB
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE | | Descriptor: | CHLORIDE ION, GLYCEROL, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based site-directed mutagenesis of hydroxynitrile lyase from cyanogenic millipede, Oxidus gracilis for enhancing catalytic efficiency and enantioselectivity
To Be Published
|
|
7YCT
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis complexed with (R)-2-Chloromandelonitrile | | Descriptor: | (2~{R})-2-(2-chlorophenyl)-2-oxidanyl-ethanenitrile, GLYCEROL, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based site-directed mutagenesis of hydroxynitrile lyase from cyanogenic millipede, Oxidus gracilis for enhancing catalytic efficiency and enantioselectivity
To Be Published
|
|
7YCD
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis bound with (R)-(+)-ALPHA-HYDROXYBENZENE-ACETONITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, Hydroxynitrile lyase, SULFATE ION | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based site-directed mutagenesis of hydroxynitrile lyase from cyanogenic millipede, Oxidus gracilis for enhancing catalytic efficiency and enantioselectivity
To Be Published
|
|
7YCF
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis IN ACETONITRILE | | Descriptor: | 2-HYDROXY-2-METHYLPROPANENITRILE, CHLORIDE ION, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based site-directed mutagenesis of hydroxynitrile lyase from cyanogenic millipede, Oxidus gracilis for enhancing catalytic efficiency and enantioselectivity
To Be Published
|
|
5XKB
 
 | | Crystal Structure of HasAp with Fe-5,15-bisethynyl-10,20-diphenylporphyrin | | Descriptor: | 5,15-Bisethynyl-10,20-diphenylporphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-05-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XIB
 
 | | Crystal Structure of HasAp with Fe-5,15-Diphenylporphyrin | | Descriptor: | 5,15-Diphenylporphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XHL
 
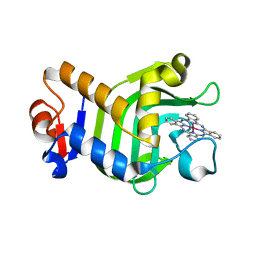 | | Crystal Structure of HasAp with Gallium Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, Phthalocyanine containing GA | | Authors: | Shoji, O, Shisaka, Y, Iwai, Y, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HasAp with Gallium Phthalocyanine
to be published
|
|
5XIE
 
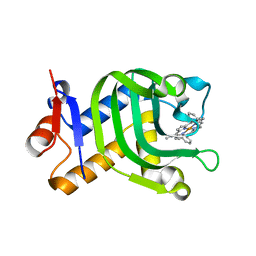 | | Crystal Structure of HasAp with 5-ethynyl-10,20-diphenylporphyrin | | Descriptor: | 5-Ethynyl-10,20-diphenylporphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2Z6B
 
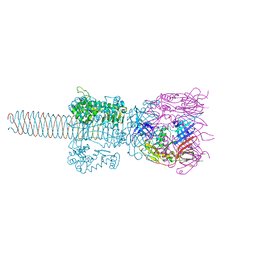 | | Crystal Structure Analysis of (gp27-gp5)3 conjugated with Fe(III) protoporphyrin | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, Baseplate structural protein Gp27, Tail-associated lysozyme | | Authors: | Koshiyama, T, Yokoi, N, Ueno, T, Kanamaru, S, Nagano, S, Shiro, Y, Arisaka, F, Watanabe, Y. | | Deposit date: | 2007-07-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor.
Small, 4, 2008
|
|
2ZQJ
 
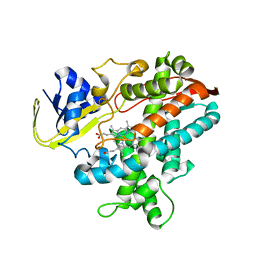 | | Substrate-Free Form of Cytochrome P450BSbeta | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-11 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
2ZQX
 
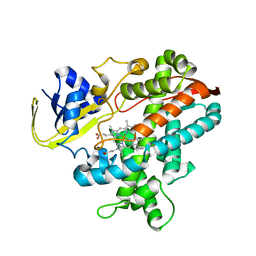 | | Cytochrome P450BSbeta cocrystallized with heptanoic acid | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
5ZLH
 
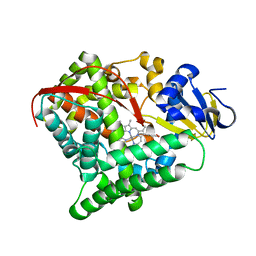 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
5ZIS
 
 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-17 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
3VOO
 
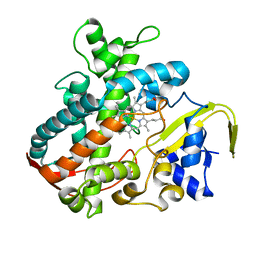 | | Cytochrome P450SP alpha (CYP152B1) mutant A245E | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3VNO
 
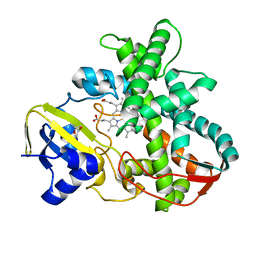 | | Cytochrome P450SP alpha (CYP152B1) mutant R241E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-01-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3VTJ
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant A245H | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3A95
 
 | | Crystal structure of hen egg white lysozyme soaked with 100mM RhCl3 at pH3.8 | | Descriptor: | CHLORIDE ION, Lysozyme C, RHODIUM(III) ION, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
3AF8
 
 | | Crystal Structure of Pd(ally)/apo-C126AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hikage, T, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanism of Accumulation and Incorporation of Organometallic Pd Complexes into the Protein Nanocage of apo-Ferritin.
Inorg.Chem., 49, 2010
|
|
