7QA2
 
 | |
7QIO
 
 | | Homology model of myosin neck domain in skeletal sarcomere | | Descriptor: | Myosin light chain 1/3, skeletal muscle isoform, Myosin regulatory light chain 2, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
7QIN
 
 | | In situ structure of actomyosin complex in skeletal sarcomere | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
7QIM
 
 | | In situ structure of nebulin bound to actin filament in skeletal sarcomere | | Descriptor: | ACTS protein, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
1OVL
 
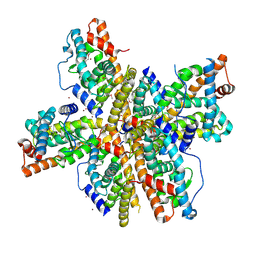 | | Crystal Structure of Nurr1 LBD | | Descriptor: | BROMIDE ION, IODIDE ION, Orphan nuclear receptor NURR1 (MSE 414, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2003-03-26 | | Release date: | 2003-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Nature, 423, 2003
|
|
4Q2J
 
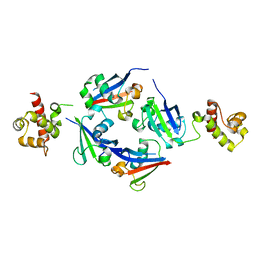 | | A novel structure-based mechanism for DNA-binding of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Wang, Z, Long, J, Yang, X, Shen, Y. | | Deposit date: | 2014-04-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Crystal structure of the ubiquitin-like domain-CUT repeat-like tandem of special AT-rich sequence binding protein 1 (SATB1) reveals a coordinating DNA-binding mechanism.
J.Biol.Chem., 289, 2014
|
|
3LQJ
 
 | |
3LQI
 
 | |
3V3L
 
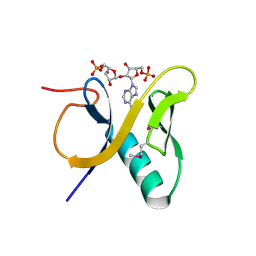 | | Crystal structure of human RNF146 WWE domain in complex with iso-ADPRibose | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146 | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Genes Dev., 26, 2012
|
|
4ERK
 
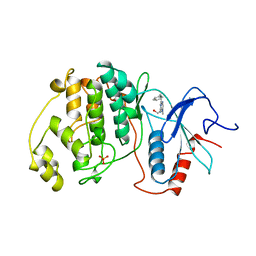 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/OLOMOUCINE | | Descriptor: | EXTRACELLULAR REGULATED KINASE 2, OLOMOUCINE, SULFATE ION | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3J42
 
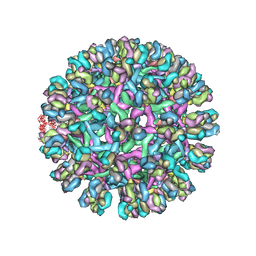 | | Obstruction of Dengue Virus Maturation by Fab Fragments of the 2H2 Antibody | | Descriptor: | Envelope protein E, Ig heavy chain V region MOPC 21, Igh protein chimera, ... | | Authors: | Wang, Z, Pennington, J.G, Jiang, W, Rossmann, M.G. | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Obstruction of Dengue Virus Maturation by Fab Fragments of the 2H2 Antibody.
J.Virol., 87, 2013
|
|
3J7L
 
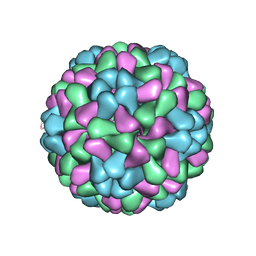 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
4FC2
 
 | |
4NA4
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4NA0
 
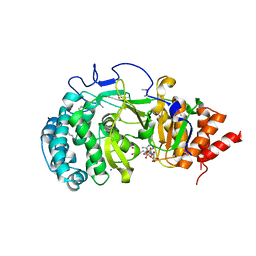 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4N9Y
 
 | |
4NA5
 
 | |
4NA6
 
 | |
4N9Z
 
 | |
3NP2
 
 | | Crystal Structure of Pd(allyl)/apo-E45C/C48A-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
3NP0
 
 | | Crystal Structure of Pd(allyl)/apo-E45C/H49A/R52H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
3NOZ
 
 | | Crystal Structure of Pd(allyl)/apo-E45C/R52H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
2FG4
 
 | | Structure of Human Ferritin L Chain | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Wang, Z, Li, C, Ellenburg, M, Ruble, J, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ferritin L chain.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
