9KT3
 
 | |
8Z6T
 
 | |
8Z6X
 
 | |
8Z6R
 
 | | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1. | | Descriptor: | CYFN1006-1 heavy chain, CYFN1006-1 light chain, Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Sun, L. | | Deposit date: | 2024-04-19 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1.
To Be Published
|
|
8Z6W
 
 | |
8Z6U
 
 | |
8Z6S
 
 | |
8Z6Q
 
 | |
8K3B
 
 | |
6K7Z
 
 | | Crystal structure of a GH18 chitinase from Pseudoalteromonas aurantia | | Descriptor: | GH18 chiitnase | | Authors: | Wang, Y.J, Li, P.Y, Cao, H.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18.
Front Microbiol, 10, 2019
|
|
8IKU
 
 | |
8J0I
 
 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
8WJW
 
 | |
8WK9
 
 | |
8WK5
 
 | |
8WK7
 
 | |
8WKA
 
 | |
7XJG
 
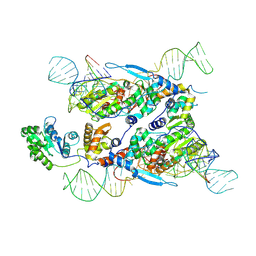 | | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | | Descriptor: | DNA (105-MER), MAGNESIUM ION, RNA (14-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2022-04-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
9JM0
 
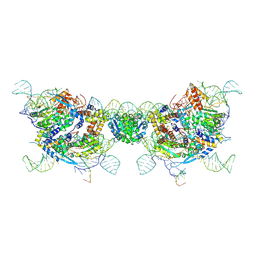 | | retron Ec86-effector fiber | | Descriptor: | DNA (85-MER), NICOTINAMIDE, RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Wang, C, Zou, T.T. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | DNA methylation activates retron Ec86 filaments for antiphage defense.
Cell Rep, 43, 2024
|
|
8YZD
 
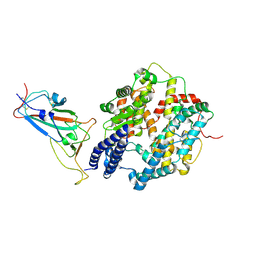 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZC
 
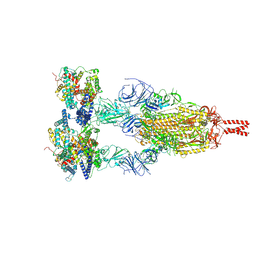 | | Structure of BA.2.86 spike protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZB
 
 | | BA.2.86 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
7V9U
 
 | | Cryo-EM structure of E.coli retron-Ec86 (RT-msDNA-RNA) at 3.2 angstrom | | Descriptor: | DNA (105-MER), RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*G)-3'), RNA (81-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
7V9X
 
 | |
