6PIQ
 
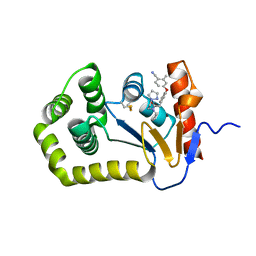 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product G6 (pyrazole 9) | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG2
 
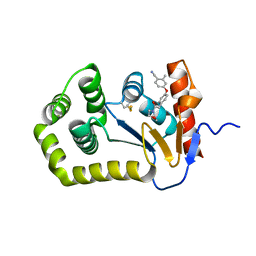 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product H5 (morpholine 8) | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PBI
 
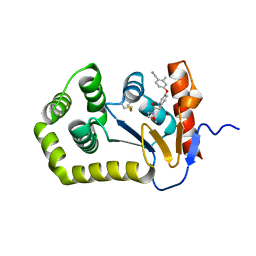 | | Crystal Structure of EcDsbA in a complex with purified morpholine 8 | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PGJ
 
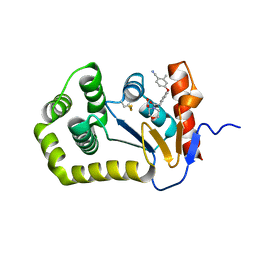 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product A5 (Morpholine carboxylic acid 7) | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, (3S)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG1
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product F1 (methylpiperazinone 6) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PDH
 
 | | Crystal Structure of EcDsbA in a complex with purified pyrazole 9 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PD7
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine carboxylic acid 7 | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-18 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
1J6T
 
 | |
8IHT
 
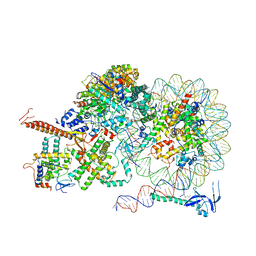 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHM
 
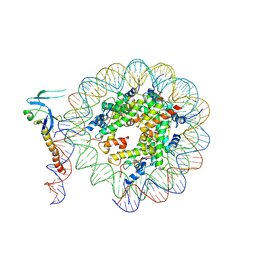 | | Eaf3 CHD domain bound to the nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHN
 
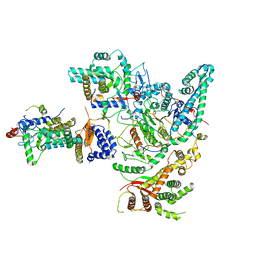 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5J8V
 
 | |
4RIE
 
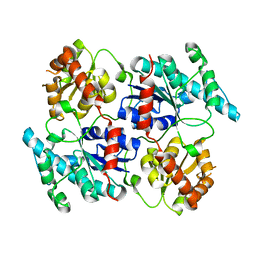 | | Landomycin Glycosyltransferase LanGT2 | | Descriptor: | Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIF
 
 | | Landomycin Glycosyltransferase LanGT2, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIG
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac | | Descriptor: | Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIH
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RII
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, TDP complex | | Descriptor: | Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RVH
 
 | | Crystal structure of MtmC in complex with SAH and TDP-4-keto-D-olivose | | Descriptor: | D-mycarose 3-C-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVD
 
 | | Crystal structure of MtmC in complex with SAM | | Descriptor: | ACETATE ION, D-mycarose 3-C-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVG
 
 | | Crystal structure of MtmC in complex with SAM and TDP | | Descriptor: | ACETATE ION, D-mycarose 3-C-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVF
 
 | | Crystal structure of MtmC in complex with TDP | | Descriptor: | D-mycarose 3-C-methyltransferase, THYMIDINE-5'-DIPHOSPHATE, ZINC ION | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RV9
 
 | | Crystal structure of MtmC in complex with SAH | | Descriptor: | ACETATE ION, CHLORIDE ION, D-mycarose 3-C-methyltransferase, ... | | Authors: | Hou, C, Chen, J.-M, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2014-11-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPP
 
 | |
