4S2H
 
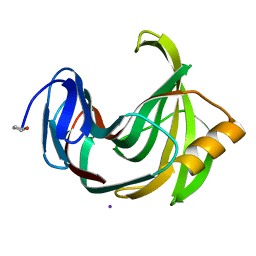 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II at pH 8.5 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Kovalevsky, A, Wan, Q, Langan, P. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2019-12-25 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4PSY
 
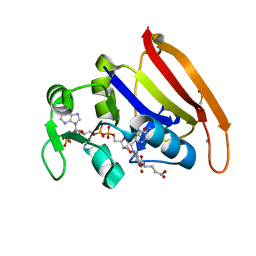 | | 100K crystal structure of Escherichia coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennet, B.C, Dealwis, C, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 22, 2014
|
|
3RSR
 
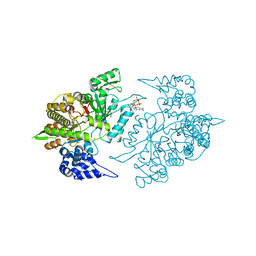 | | Crystal Structure of 5-NITP Inhibition of Yeast Ribonucleotide Reductase | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO-1H-INDOLE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Wan, Q, Mohammed, F, Jha, S, Motea, E, Berdis, A, Dealwis, C.G. | | Deposit date: | 2011-05-02 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evaluating the therapeutic potential of a non-natural nucleotide that inhibits human ribonucleotide reductase.
Mol.Cancer Ther., 11, 2012
|
|
6JXL
 
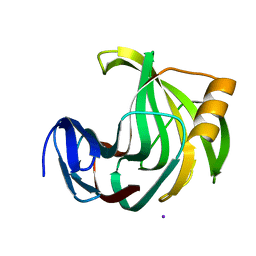 | |
7EO6
 
 | | X-ray structure analysis of xylanase | | Descriptor: | Endo-1,4-beta-xylanase, IODIDE ION | | Authors: | Wan, Q, Yi, Y, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and structural analysis of a thermophilic GH11 xylanase from compost metatranscriptome.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
7EO3
 
 | | X-ray structure analysis of beita-1,3-glucanase | | Descriptor: | 1,3-beta-glucanase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION | | Authors: | Wan, Q, Feng, J, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.141 Å) | | Cite: | Identification and structural analysis of a thermophilic beta-1,3-glucanase from compost
Bioresour Bioprocess, 8, 2021
|
|
6JZP
 
 | |
6K9X
 
 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-18 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism
Acta Crystallographica Section D-Biological Crystallography, D70, 2014
|
|
6KVV
 
 | |
6KWE
 
 | |
6KWH
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism
To Be Published
|
|
3EL2
 
 | | Crystal Structure of Monomeric Actin Bound to Ca-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
3EKU
 
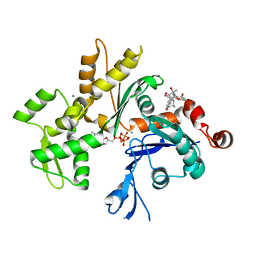 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
3EKS
 
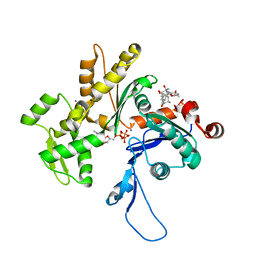 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
2HF4
 
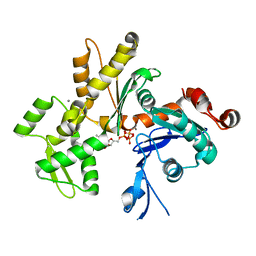 | | Crystal structure of Monomeric Actin in its ATP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Rould, M.A, Wan, Q, Joel, P.B, Lowey, S, Trybus, K.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Expressed Non-polymerizable Monomeric Actin in the ADP and ATP States.
J.Biol.Chem., 281, 2006
|
|
2HF3
 
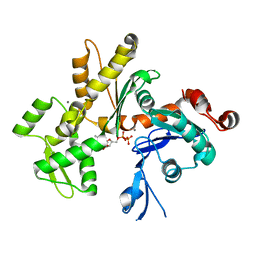 | | Crystal structure of monomeric Actin in the ADP bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Rould, M.A, Wan, Q, Joel, P.B, Lowey, S, Trybus, K.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Expressed Non-polymerizable Monomeric Actin in the ADP and ATP States.
J.Biol.Chem., 281, 2006
|
|
3WP1
 
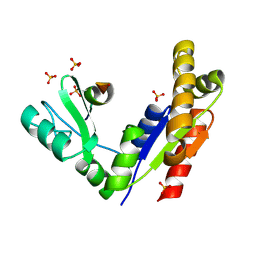 | | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl | | Descriptor: | Disks large homolog 4, Lethal(2) giant larvae protein homolog 2, SULFATE ION | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
3WP0
 
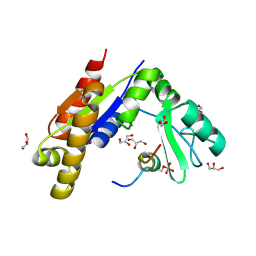 | | Crystal structure of Dlg GK in complex with a phosphor-Lgl2 peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, Lethal(2) giant larvae protein homolog 2 | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
3S8B
 
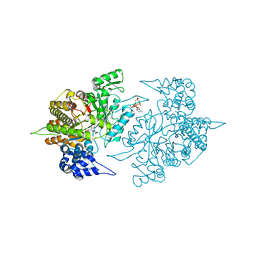 | | Structure of Yeast Ribonucleotide Reductase 1 with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3S8A
 
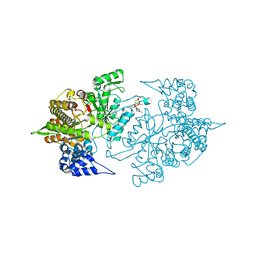 | | Structure of Yeast Ribonucleotide Reductase R293A with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3S87
 
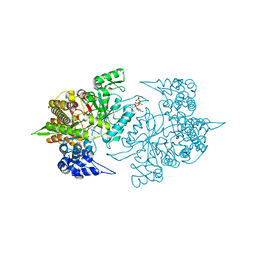 | | Structure of Yeast Ribonucleotide Reductase 1 with dGTP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3S8C
 
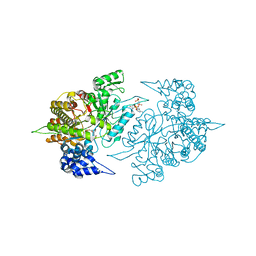 | | Structure of Yeast Ribonucleotide Reductase 1 R293A with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3TBA
 
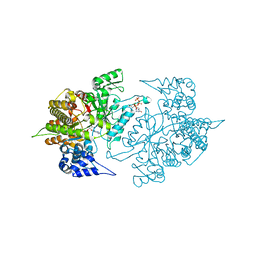 | | Structure of Yeast Ribonucleotide Reductase 1 Q288A with dGTP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C. | | Deposit date: | 2011-08-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of Arginine 293 and Glutamine 288 in Communication between Catalytic and Allosteric Sites in Yeast Ribonucleotide Reductase.
J.Mol.Biol., 419, 2012
|
|
3TB9
 
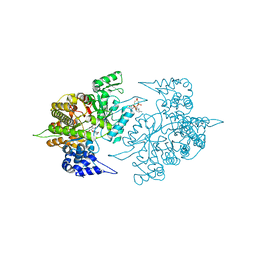 | | Structure of Yeast Ribonucleotide Reductase 1 Q288A with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-08-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of Arginine 293 and Glutamine 288 in Communication between Catalytic and Allosteric Sites in Yeast Ribonucleotide Reductase.
J.Mol.Biol., 419, 2012
|
|
6JWB
 
 | | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
