5DPT
 
 | | Crystal structure of PLEKHM1 LIR-fused human GABARAPL1_2-117 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Pleckstrin homology domain-containing family M member 1, Gamma-aminobutyric acid receptor-associated protein-like 1,Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Ravichandran, A.C, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional analysis of the GABARAP interaction motif (GIM).
EMBO Rep., 18, 2017
|
|
5DPS
 
 | |
5DPW
 
 | |
5DPR
 
 | |
8B01
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc | | Descriptor: | DECANE, DOCOSANE, HEXANE, ... | | Authors: | Davies, J.S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2022-09-06 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
6VJR
 
 | |
4FWG
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with lactacystin | | Descriptor: | Omuralide, open form, PHOSPHATE ION, ... | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FWD
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FWH
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with MG262 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G01
 
 | | ARA7-GDP-Ca2+/VPS9a | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein RABF2b, ... | | Authors: | Ihara, K. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct metal recognition by guanine nucleotide-exchange factor in the initial step of the exchange reaction
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7XPD
 
 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPE
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPF
 
 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPB
 
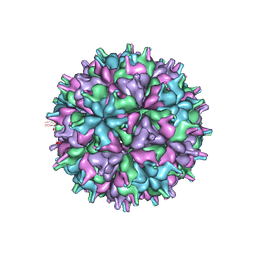 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPA
 
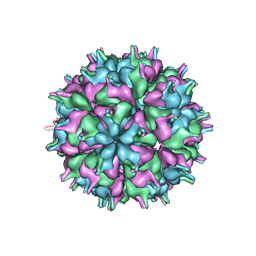 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XGZ
 
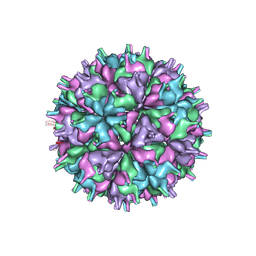 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPG
 
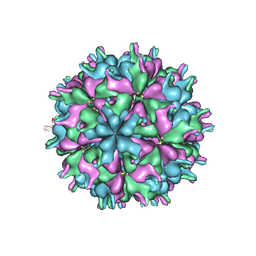 | | Cryo-EM structure of the T=3 lake sinai virus 1 (delta-N48) virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha, RNA (5'-R(P*UP*G)-3') | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
1HUS
 
 | | RIBOSOMAL PROTEIN S7 | | Descriptor: | RIBOSOMAL PROTEIN S7 | | Authors: | Hosaka, H, Nakagawa, A, Tanaka, I. | | Deposit date: | 1997-08-08 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor.
Structure, 5, 1997
|
|
1VIN
 
 | |
1J1E
 
 | | Crystal structure of the 52kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
1J1D
 
 | | Crystal structure of the 46kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
2EIR
 
 | |
2EIO
 
 | |
2EIQ
 
 | |
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
