7R9B
 
 | |
7QWE
 
 | |
7QWB
 
 | |
7QWA
 
 | |
7QWD
 
 | |
7QWC
 
 | |
7RGC
 
 | |
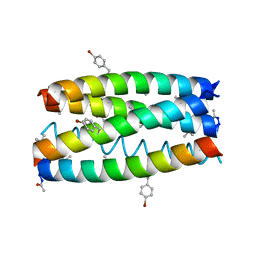
7RPP
 
 | | Crystal structure of human CEACAM1 with GFCC' and ABED face | | Descriptor: | 1,2-ETHANEDIOL, Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of human CEACAM1 oligomerization.
Commun Biol, 5, 2022
|
|
7RMN
 
 | |
7RPN
 
 | |
7RCQ
 
 | |
7SPY
 
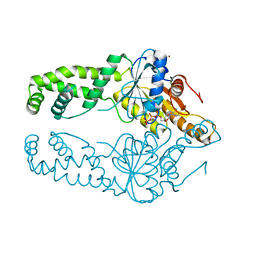 | | Get3 bound to ATP from G. intestinalis in the closed form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SQ0
 
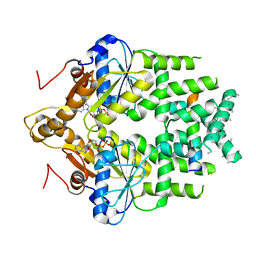 | | Get3 bound to ADP and the transmembrane domain of the tail-anchored protein Bos1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase ASNA1 homolog, MAGNESIUM ION, ... | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SPZ
 
 | | Nucleotide-free Get3 in two open forms | | Descriptor: | ATPase ASNA1 homolog, ZINC ION | | Authors: | Fry, M.Y, Maggiolo, A.O, Clemons Jr, W.M. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2M9U
 
 | | Solution NMR structure of the C-terminal domain (CTD) of Moloney murine leukemia virus integrase, Northeast Structural Genomics Target OR41A | | Descriptor: | Integrase p46 | | Authors: | Aiyer, S, Rossi, P, Schneider, W.M, Chander, A, Roth, M.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction.
Nucleic Acids Res., 42, 2014
|
|
2MK0
 
 | | Structure of the PSCD4-domain of the cell wall protein pleuralin-1 from the diatom Cylindrotheca fusiformis | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Deutzmann, R, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, S.H.R. | | Deposit date: | 2014-01-22 | | Release date: | 2015-02-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2MZB
 
 | | Solution structural studies of GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) from Methanocaldococcus jannaschii | | Descriptor: | Adenosylcobinamide-phosphate guanylyltransferase | | Authors: | Singarapu, K, Otte, M, Tonelli, M, Westler, W.M, Escalante-Semerena, J.C, Markley, J.L. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Studies of GTP:Adenosylcobinamide-Phosphateguanylyl Transferase (CobY) from Methanocaldococcus jannaschii.
Plos One, 10, 2015
|
|
2N08
 
 | | NMR structure of a short hydrophobic 11mer peptide in 25 mM SDS solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N0I
 
 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N09
 
 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2NBI
 
 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
2NZT
 
 | | Crystal structure of human hexokinase II | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Hexokinase-2, UNKNOWN ATOM OR ION, ... | | Authors: | Rabeh, W.M, Zhu, H, Nedyalkova, L, Tempel, W, Wasney, G, Landry, R, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-25 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation.
Biosci.Rep., 38, 2018
|
|
2OVN
 
 | |
2P0E
 
 | | Human nicotinamide riboside kinase 1 in complex with tiazofurin | | Descriptor: | (1R)-1-[4-(AMINOCARBONYL)-1,3-THIAZOL-2-YL]-1,4-ANHYDRO-D-RIBITOL, CHLORIDE ION, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
