379D
 
 | | THE STRUCTURAL BASIS OF HAMMERHEAD RIBOZYME SELF-CLEAVAGE | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*CP*UP*CP*GP*UP*AP*AP*GP*A P*GP*UP*CP*AP*CP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3') | | Authors: | Murray, J.B, Terwey, D.P, Maloney, L, Karpeisky, A, Usman, N, Beigelman, L, Scott, W.G. | | Deposit date: | 1998-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of hammerhead ribozyme self-cleavage.
Cell(Cambridge,Mass.), 92, 1998
|
|
6NCY
 
 | | Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans | | Descriptor: | Beta-glucuronidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selecting a Single Stereocenter: The Molecular Nuances That Differentiate beta-Hexuronidases in the Human Gut Microbiome.
Biochemistry, 58, 2019
|
|
6NCW
 
 | |
6NCX
 
 | |
300D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | Descriptor: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
3BSQ
 
 | | Crystal structure of human kallikrein 7 produced as a secretion protein in E.coli | | Descriptor: | Kallikrein-7, SULFATE ION | | Authors: | Fernandez, I.S, Standker, L, Magert, H.J, Forssmann, W.G, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human epidermal kallikrein 7 (hK7) synthesized directly in its native state in E. coli: insights into the atomic basis of its inhibition by LEKTI domain 6 (LD6)
J.Mol.Biol., 377, 2008
|
|
2GQ2
 
 | | Mycobacterium tuberculosis ThyX-NADP complex | | Descriptor: | GLYCEROL, IODIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sampathkumar, P, Turley, S, Sibley, C.H, Hol, W.G. | | Deposit date: | 2006-04-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NADP+ expels both the co-factor and a substrate analog from the Mycobacterium tuberculosis ThyX active site: opportunities for anti-bacterial drug design.
J.Mol.Biol., 360, 2006
|
|
2HCC
 
 | | SOLUTION STRUCTURE OF THE HUMAN CHEMOKINE HCC-2, NMR, 30 STRUCTURES | | Descriptor: | HUMAN CHEMOKINE HCC-2 | | Authors: | Sticht, H, Escher, S.E, Schweimer, K, Forssmann, W.G, Roesch, P, Adermann, K. | | Deposit date: | 1998-07-03 | | Release date: | 1999-07-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype.
Biochemistry, 38, 1999
|
|
4NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
8DTQ
 
 | | Crystal Structure of Staphylococcus aureus pSK41 Cop | | Descriptor: | CHLORIDE ION, Helix-turn-helix domain, SODIUM ION | | Authors: | Walton, W.G, Eakes, T.C, Redinbo, M.R, McLaughlin, K.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | pSK41/pGO1-family conjugative plasmids of Staphylococcus aureus encode a cryptic repressor of replication.
Plasmid, 128, 2023
|
|
4FUL
 
 | | PI3 Kinase Gamma bound to a pyrmidine inhibitor | | Descriptor: | 4-({4-[3-(piperidin-1-ylcarbonyl)phenyl]pyrimidin-2-yl}amino)benzenesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Gopalsamy, A, Bennett, E.M, Shi, M, Zhang, W.G, Bard, J, Yu, K. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-17 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification of pyrimidine derivatives as hSMG-1 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6AZH
 
 | |
1NNA
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1NNB
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1DUH
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
1E4T
 
 | | Solution structure of the mouse defensin mBD-7 | | Descriptor: | Beta-defensin 7 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1E4Q
 
 | | Solution structure of the human defensin hBD-2 | | Descriptor: | BETA-DEFENSIN 2 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
1E4R
 
 | | Solution structure of the mouse defensin mBD-8 | | Descriptor: | Beta-defensin 8 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1EJ9
 
 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
1E4S
 
 | | Solution structure of the human defensin hBD-1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
5COR
 
 | |
2ISY
 
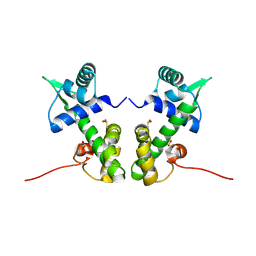 | | Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR) | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2ISZ
 
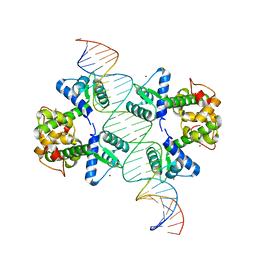 | | Crystal structure of a two-domain IdeR-DNA complex crystal form I | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2IT0
 
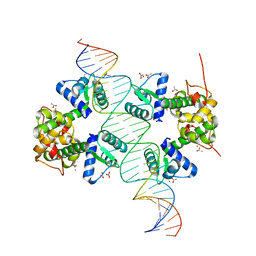 | | Crystal structure of a two-domain IdeR-DNA complex crystal form II | | Descriptor: | ACETATE ION, Iron-dependent repressor ideR, NICKEL (II) ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2LT9
 
 | |
