4YQJ
 
 | |
4YQR
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)(4-methylpiperazin-1-yl)methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
4YPY
 
 | |
4YQ5
 
 | |
4YQG
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-(cyclohexylmethyl)-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Williams, S.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
4YQO
 
 | |
4YPZ
 
 | |
4YQ4
 
 | |
4YQD
 
 | |
1JI4
 
 | | NAP protein from helicobacter pylori | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, NEUTROPHIL-ACTIVATING PROTEIN A, ... | | Authors: | Zanotti, G, Papinutto, E, Dundon, W.G, Battistutta, R, Seveso, M, Del Giudice, G, Rappuoli, R, Montecucco, C. | | Deposit date: | 2001-06-29 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Neutrophil-activating Protein from Helicobacter pylori
J.Mol.Biol., 323, 2002
|
|
1JI5
 
 | | Dlp-1 from bacillus anthracis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dlp-1, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
1JIG
 
 | | Dlp-2 from Bacillus anthracis | | Descriptor: | Dlp-2, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
1LM8
 
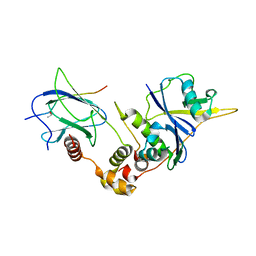 | | Structure of a HIF-1a-pVHL-ElonginB-ElonginC Complex | | Descriptor: | ELONGIN B, ELONGIN C, Hypoxia-inducible factor 1 alpha, ... | | Authors: | Min, J.-H, Yang, H, Ivan, M, Gertler, F, Kaelin JR, W.G, Pavletich, N.P. | | Deposit date: | 2002-04-30 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling.
Science, 296, 2002
|
|
1MME
 
 | |
1N1G
 
 | |
1NYI
 
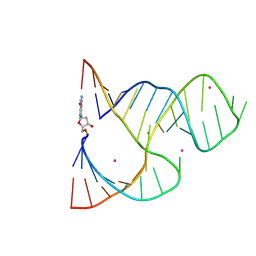 | | Crosslinked Hammerhead Ribozyme Initial State | | Descriptor: | 5'-R(*GP*CP*CP*GP*AP*AP*AP*CP*UP*CP*GP*UP*AP*AP*GP*AP*GP*UP*CP*AP*CP*CP*AP*C)-3', 5'-R(P*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3', COBALT (II) ION, ... | | Authors: | Dunham, C.M, Murray, J.B, Scott, W.G. | | Deposit date: | 2003-02-12 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme.
J.Mol.Biol., 332, 2003
|
|
1GNH
 
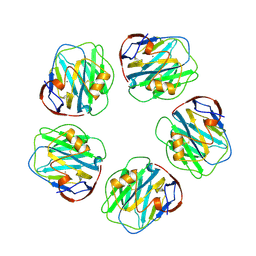 | | HUMAN C-REACTIVE PROTEIN | | Descriptor: | C-REACTIVE PROTEIN, CALCIUM ION | | Authors: | Shrive, A.K, Cheetham, G.M.T, Holden, D, Myles, D.A, Turnell, W.G, Volanakis, J.E, Pepys, M.B, Bloomer, A.C, Greenhough, T.J. | | Deposit date: | 1996-03-01 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three dimensional structure of human C-reactive protein.
Nat.Struct.Biol., 3, 1996
|
|
1GAC
 
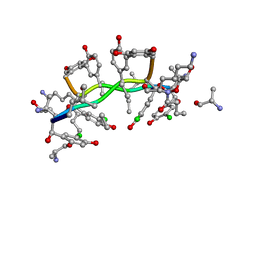 | | NMR structure of asymmetric homodimer of a82846b, a glycopeptide antibiotic, complexed with its cell wall pentapeptide fragment | | Descriptor: | CELL WALL PENTAPEPTIDE, CHLOROORIENTICIN A, vancosamine, ... | | Authors: | Kline, A.D, Prowse, W.G, Skelton, M.A, Loncharich, R.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformation of A82846B, a Glycopeptide Antibiotic, Complexed with its Cell Wall Fragment: An Asymmetric Homodimer Determined Using NMR Spectroscopy.
Biochemistry, 34, 1995
|
|
1IDE
 
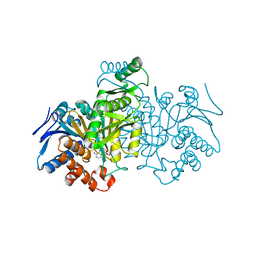 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDC
 
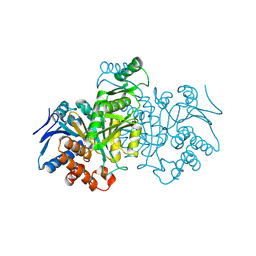 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDF
 
 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
7DT0
 
 | | Proline hydroxylase H11-N101I mutant | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.G, Yang, L.Y. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline
To Be Published
|
|
7E01
 
 | |
7E05
 
 | |
7E08
 
 | |
