5OR5
 
 | |
5IEB
 
 | |
4LUP
 
 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1LUA
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 complexed with NADP | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
1LU9
 
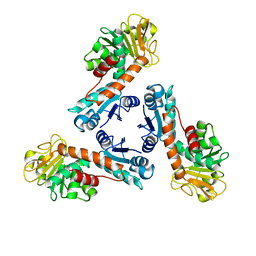 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
5IEJ
 
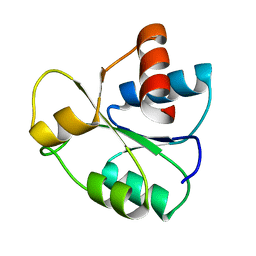 | |
1XA8
 
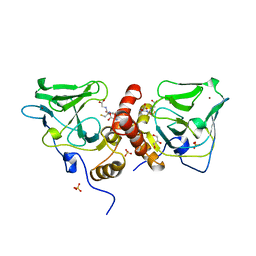 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1X6M
 
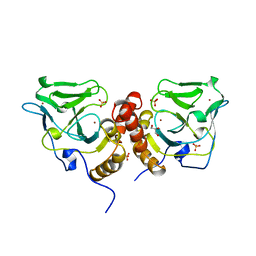 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
2LFW
 
 | | NMR structure of the PhyRSL-NepR complex from Sphingomonas sp. Fr1 | | Descriptor: | NepR anti sigma factor, PhyR sigma-like domain | | Authors: | Campagne, S, Damberger, F.F, Vorholt, J.A, Allain, F.H.-T. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MAO
 
 | |
2MAP
 
 | |
1Y5Y
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 | | Descriptor: | CALCIUM ION, Formaldehyde-activating enzyme fae, SODIUM ION | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
1Y60
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 with bound 5,10-methylene tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Formaldehyde-activating enzyme fae | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
6S6Y
 
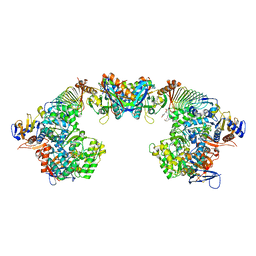 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | Descriptor: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | Authors: | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4WAS
 
 | | STRUCTURE OF THE ETR1P/NADP/CROTONYL-COA COMPLEX | | Descriptor: | CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Capitani, G, Erb, T.J. | | Deposit date: | 2014-08-31 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
4W99
 
 | | Apo-structure of the Y79F,W322E-double mutant of Etr1p | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Erb, T.J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
6GS8
 
 | | Crystal structure of SmbA in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Uncharacterized protein | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2018-06-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus
Nat Microbiol, 2021
|
|
6GTM
 
 | | Crystal structure of SmbA in complex with ppGpp. | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, SmbA | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus
Nat Microbiol, 2021
|
|
1QLM
 
 | |
4F18
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 8.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
4F19
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 4.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
4F1U
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 4.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, ... | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
4F1V
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 8.5 | | Descriptor: | HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, SULFATE ION | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
