7UWZ
 
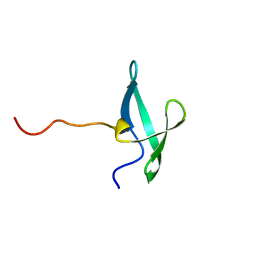 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 33_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 29_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1XFL
 
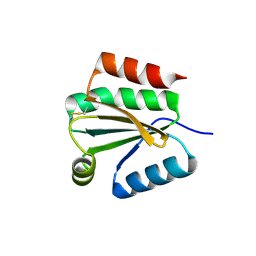 | | Solution Structure of Thioredoxin h1 from Arabidopsis Thaliana | | Descriptor: | Thioredoxin h1 | | Authors: | Peterson, F.C, Lytle, B.L, Sampath, S, Vinarov, D, Tyler, E, Shahan, M, Markley, J.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thioredoxin h1 from Arabidopsis thaliana.
Protein Sci., 14, 2005
|
|
1YEL
 
 | | Structure of the hypothetical Arabidopsis thaliana protein At1g16640.1 | | Descriptor: | At1g16640 | | Authors: | Peterson, F.C, Waltner, J.K, Lytle, B.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the B3 domain from Arabidopsis thaliana protein At1g16640
Protein Sci., 14, 2005
|
|
1ZR9
 
 | |
2B3I
 
 | | NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PROKARYOTE, PROCHLOROTHRIX HOLLANDICA (19 STRUCTURES) | | Descriptor: | COPPER (I) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Babu, C.R, Volkman, B.F, Bullerjahn, G.S. | | Deposit date: | 1998-12-11 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of plastocyanin from the photosynthetic prokaryote, Prochlorothrix hollandica.
Biochemistry, 38, 1999
|
|
2HDL
 
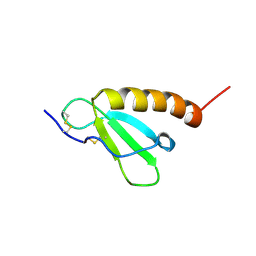 | | Solution structure of Brak/CXCL14 | | Descriptor: | Small inducible cytokine B14 | | Authors: | Peterson, F.C, Thorpe, J.A, Harder, A.G, Volkman, B.F, Schwarze, S.R. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants Involved in the Regulation of CXCL14/BRAK Expression by the 26 S Proteasome.
J.Mol.Biol., 363, 2006
|
|
1DAV
 
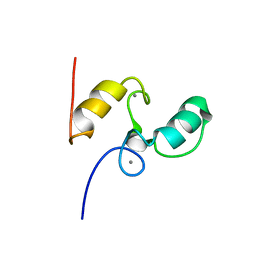 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (20 STRUCTURES) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1DAQ
 
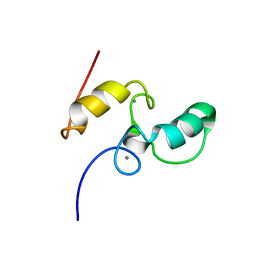 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1DC7
 
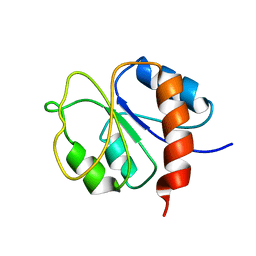 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
1DC8
 
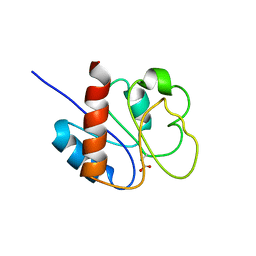 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
3NJ0
 
 | | X-ray crystal structure of the PYL2-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJO
 
 | | X-ray crystal structure of the Pyr1-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYR1, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Peterson, F.C, Volkman, B.F, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJ1
 
 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5UR5
 
 | | PYR1 bound to the rationally designed agonist 4m | | Descriptor: | Abscisic acid receptor PYR1, GLYCEROL, N-(4-cyano-3-ethyl-5-methylphenyl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5UR6
 
 | | PYR1 bound to the rationally designed agonist cyanabactin | | Descriptor: | Abscisic acid receptor PYR1, N-(4-cyano-3-cyclopropylphenyl)-1-(4-methylphenyl)methanesulfonamide, SULFATE ION | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5UR7
 
 | |
5US5
 
 | | Solution structure of the IreB homodimer | | Descriptor: | UPF0297 protein EF_1202 | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Kristich, C.J, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2017-02-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dimerization of IreB, a Negative Regulator of Cephalosporin Resistance in Enterococcus faecalis.
J. Mol. Biol., 429, 2017
|
|
1Q53
 
 | |
1PUX
 
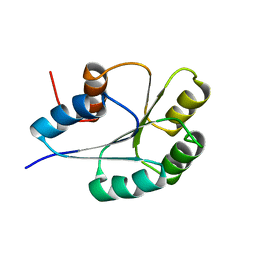 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
1J8I
 
 | | Solution Structure of Human Lymphotactin | | Descriptor: | Lymphotactin | | Authors: | Kuloglu, E.S, McCaslin, D.R, Markley, J.L, Pauza, C.D, Volkman, B.F. | | Deposit date: | 2001-05-21 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
1J9O
 
 | | SOLUTION STRUCTURE OF HUMAN LYMPHOTACTIN | | Descriptor: | LYMPHOTACTIN | | Authors: | Kuloglu, E.S, McCaslin, D.R, Kitabwalla, M, Pauza, C.D, Markley, J.L, Volkman, B.F. | | Deposit date: | 2001-05-28 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
1JJZ
 
 | |
1K48
 
 | |
1RY4
 
 | |
