3BRO
 
 | | Crystal structure of the transcription regulator MarR from Oenococcus oeni PSU-1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | Authors: | Kim, Y, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Transcription Regulator MarR from Oenococcus oeni PSU-1.
To be Published
|
|
2QZG
 
 | | Crystal structure of unknown function protein MMP1188 | | Descriptor: | Conserved uncharacterized archaeal protein | | Authors: | Chang, C, Perez, V, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MMP1188, unknown function protein.
To be Published
|
|
2QHQ
 
 | | Crystal structure of unknown function protein VPA0580 | | Descriptor: | ACETATE ION, Unknown function protein VPA0580 | | Authors: | Chang, C, Kim, Y, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of unknown function protein VPA0580.
To be Published
|
|
2R8D
 
 | |
2B67
 
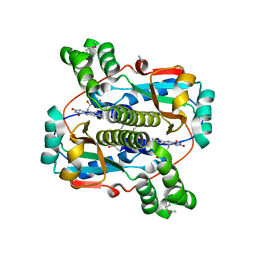 | | Crystal structure of the Nitroreductase Family Protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | ACETIC ACID, COG0778: Nitroreductase, FLAVIN MONONUCLEOTIDE | | Authors: | Kim, Y, Volkart, L, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Nitroreductase Family Protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
2GKP
 
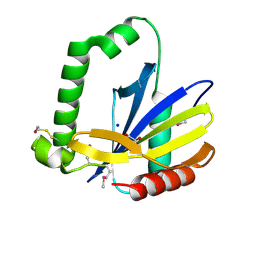 | | Protein of Unknown Function NMB0488 from Neisseria meningitidis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Osipiuk, J, Volkart, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-03 | | Release date: | 2006-05-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray crystal structure of hypothetical protein NMB0488 from Neisseria meningitidis.
To be Published
|
|
2GMQ
 
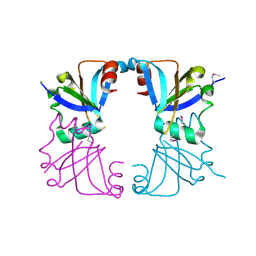 | |
2F06
 
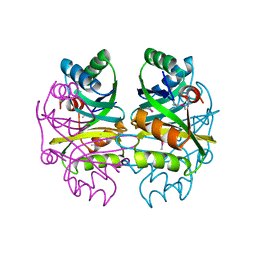 | |
2G7Z
 
 | | Conserved DegV-like Protein of Unknown Function from Streptococcus pyogenes M1 GAS Binds Long-chain Fatty Acids | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, ... | | Authors: | Nocek, B, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conserved hypothetical protein from Streptococcus pyogenes M1 GAS discloses long-fatty acid (heptadecanoic acid) binding function
To be Published
|
|
3BJ6
 
 | | Crystal structure of MarR family transcription regulator SP03579 | | Descriptor: | ETHANOL, Transcriptional regulator, MarR family | | Authors: | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of MarR family Transcription Regulator SP03579.
To be Published
|
|
3BOQ
 
 | |
3GNJ
 
 | | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB | | Descriptor: | Thioredoxin domain protein | | Authors: | Tan, K, Volkart, L, Gu, M, Kinney, J.N, Babnigg, G, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB
To be Published
|
|
3K2V
 
 | | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae. | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae.
TO BE PUBLISHED
|
|
3JRK
 
 | | A putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes | | Descriptor: | GLYCEROL, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | Fan, Y, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of a putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes
To be Published
|
|
3JYF
 
 | | The crystal structure of a 2,3-cyclic nucleotide 2-phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor protein from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fan, Y, Volkart, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-21 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a 2,3-cyclic nucleotide 2-phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor protein from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3OVK
 
 | |
3OHR
 
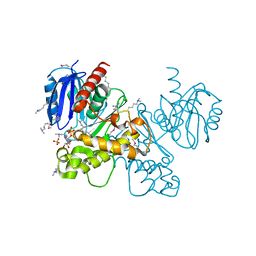 | | Crystal structure of fructokinase from bacillus subtilis complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative fructokinase, SULFATE ION, ... | | Authors: | Nocek, B, Volkart, L, Cuff, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
3ONO
 
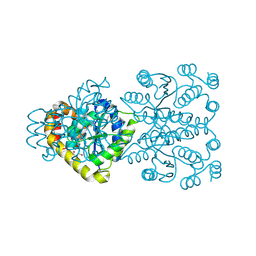 | |
3NWG
 
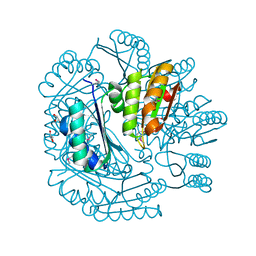 | | The crystal structure of a microcomparments protein from Desulfitobacterium hafniense DCB | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Fan, Y, Volkart, L, Gu, M, Axen, S, Greenleaf, W.B, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-09 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a PduT homolog from a novel bacterial microcompartment
To be Published
|
|
3CNE
 
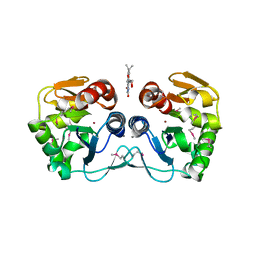 | | Crystal structure of the putative protease I from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Putative protease I, ... | | Authors: | Zhang, R, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the putative protease I from Bacteroides thetaiotaomicron.
To be Published
|
|
3N70
 
 | | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A | | Descriptor: | SULFATE ION, Transport activator | | Authors: | Stein, A.J, Mulligan, R, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A
To be Published
|
|
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3BY6
 
 | |
3CDH
 
 | | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi DSS-3 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi.
To be Published
|
|
3EEH
 
 | |
