5HCW
 
 | |
5V0F
 
 | | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone | | 分子名称: | 4-arsanyl-2-nitrophenol, CHLORIDE ION, FE (III) ION, ... | | 著者 | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | 登録日 | 2017-02-28 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone.
To Be Published
|
|
3E9W
 
 | |
3Q5C
 
 | |
3QK4
 
 | |
5C68
 
 | |
5C4P
 
 | |
5C6X
 
 | | Crystal structure of C-As lyase with Co(II) | | 分子名称: | COBALT (II) ION, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | 著者 | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | 登録日 | 2015-06-23 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of C-As lyase with Co(II)
To Be Published
|
|
5CB9
 
 | | Crystal structure of C-As lyase with mercaptoethonal | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | 著者 | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | 登録日 | 2015-06-30 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
5D4F
 
 | | Crystal structure of C-As lyase with Fe(III) | | 分子名称: | CHLORIDE ION, FE (III) ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | 著者 | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | 登録日 | 2015-08-07 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
5DFG
 
 | |
5DRH
 
 | | Crystal structure of apo C-As lyase | | 分子名称: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | 著者 | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | 登録日 | 2015-09-15 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal structure of apo C-As lyase
To Be Published
|
|
3FQ5
 
 | |
3G2R
 
 | |
3G2A
 
 | |
3HQE
 
 | |
3HS1
 
 | |
3IXN
 
 | |
3R86
 
 | |
3TCI
 
 | |
7RST
 
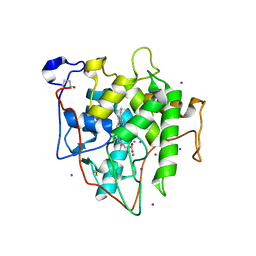 | | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chloroperoxidase, ... | | 著者 | Tang, X, Venkadesh, S, Zhou, J, Rosen, B, Wang, X. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger
To Be Published
|
|
4KSM
 
 | |
3F8O
 
 | |
4KX4
 
 | |
4DWY
 
 | | Interactions of Mn2+ with a non-self-complementary Z-type DNA duplex | | 分子名称: | DNA (5'-D(*CP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*G)-3'), MANGANESE (II) ION | | 著者 | Mandal, P.K, Venkadesh, S, Gautham, N. | | 登録日 | 2012-02-27 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Interactions of Mn(2+) with a non-self-complementary Z-type DNA duplex
Acta Crystallogr.,Sect.F, 68, 2012
|
|
