1USY
 
 | | ATP phosphoribosyl transferase (HisG:HisZ) complex from Thermotoga maritima | | Descriptor: | ATP PHOSPHORIBOSYLTRANSFERASE, ATP PHOSPHORIBOSYLTRANSFERASE REGULATORY SUBUNIT, HISTIDINE, ... | | Authors: | Vega, M.C, Fernandez, F.J, Murphy, G.E, Zou, P, Popov, A, Wilmanns, M. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Regulation of the Hetero-Octameric ATP Phosphoribosyl Transferase Complex from Thermotoga Maritima by a tRNA Synthetase-Like Subunit.
Mol.Microbiol., 55, 2005
|
|
264D
 
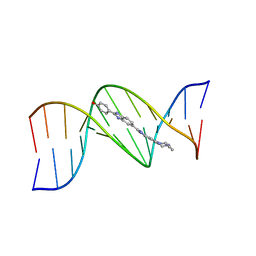 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR-GROOVE-BINDING DRUG HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Vega, M.C, Garcia-Saez, I, Aymami, J, Eritja, R, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Coll, M. | | Deposit date: | 1994-09-22 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258.
Eur.J.Biochem., 222, 1994
|
|
1BAY
 
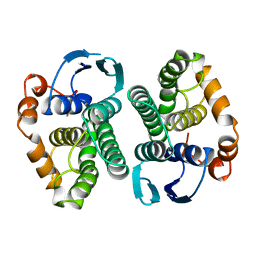 | | GLUTATHIONE S-TRANSFERASE YFYF CYS 47-CARBOXYMETHYLATED CLASS PI, FREE ENZYME | | Descriptor: | GLUTATHIONE S-TRANSFERASE CLASS PI | | Authors: | Vega, M.C, Coll, M. | | Deposit date: | 1996-11-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
1QKW
 
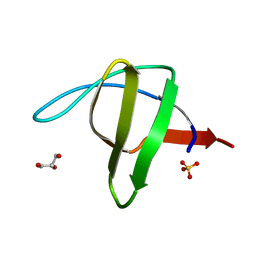 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
1QKX
 
 | |
1GTI
 
 | | MODIFIED GLUTATHIONE S-TRANSFERASE (PI) COMPLEXED WITH S (P-NITROBENZYL)GLUTATHIONE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, S-(P-NITROBENZYL)GLUTATHIONE | | Authors: | Vega, M.C, Coll, M. | | Deposit date: | 1998-01-09 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
1E6H
 
 | |
2WJZ
 
 | | Crystal structure of (HisH) K181A Y138A mutant of imidazoleglycerolphosphate synthase (HisH HisF) which displays constitutive glutaminase activity | | Descriptor: | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE HISF, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISH, PHOSPHATE ION | | Authors: | Vega, M.C, List, F, Razeto, A, Haeger, M.C, Babinger, K, Kuper, J, Sterner, R, Wilmanns, M. | | Deposit date: | 2009-06-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
1WAA
 
 | | IG27 protein domain | | Descriptor: | TITIN, ZINC ION | | Authors: | Vega, M.C, Valencia, L, Zou, P, Wilmanns, M. | | Deposit date: | 2004-10-25 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanical Network in Titin Immunoglobulin from Force Distribution Analysis.
Plos Comput.Biol., 5, 2009
|
|
1UU1
 
 | | Complex of Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER | | Authors: | Vega, M.C, Fernandez, F.J, Lehman, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU2
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (apo-form) | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-13 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU0
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UUE
 
 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3ZR4
 
 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
1E6G
 
 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25I, V53I, V58L MUTANT | | Descriptor: | SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-15 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Strain in the Hydrophobic Core and its Implications for Protein Folding and Design
Nat.Struct.Biol., 9, 2002
|
|
1E7O
 
 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25V, V44I, V58L MUTATIONS | | Descriptor: | GLYCEROL, SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-31 | | Release date: | 2003-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Thermodynamic and Kinetic Analysis of the Folding Pathway of an SH3 Domain Entropically Stabilised by a Redesigned Hydrophobic Core
J.Mol.Biol., 328, 2003
|
|
1H8K
 
 | |
1H1C
 
 | |
6TJP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve - P212121 | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Fernandez, F.J, Machon, C, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5NQ6
 
 | | Crystal structure of the inhibited form of the redox-sensitive SufE-like sulfur acceptor CsdE from Escherichia coli at 2.40 Angstrom Resolution | | Descriptor: | GLYCEROL, SULFATE ION, Sulfur acceptor protein CsdE | | Authors: | Penya-Soler, E, Aranda, J, Lopez-Estepa, M, Gomez, S, Garces, F, Coll, M, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the inhibited form of the redox-sensitive SufE-like sulfur acceptor CsdE.
PLoS ONE, 12, 2017
|
|
5O5Z
 
 | | CRYSTAL STRUCTURE OF THERMOCOCCUS LITORALIS ADP-DEPENDENT GLUCOKINASE (GK) | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5Y
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5X
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-28 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
8OHA
 
 | | Crystal structure of Leptospira interrogans GAPDH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Navas-Yuste, S, de la Paz, K, Querol-Garcia, J, Gomez-Quevedo, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity.
Front Immunol, 14, 2023
|
|
