8Q40
 
 | | Crystal structure of cA4 activated Can2 in complex with a cleaved DNA substrate | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*CP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q42
 
 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-A DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*AP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q41
 
 | | Crystal structure of Can2 (E341A) bound to cA4 and TTTAAA ssDNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q43
 
 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-C DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*CP*CP*CP*CP*C)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q44
 
 | | Crystal structure of cA4-bound Can2 (E364R) in complex with oligo-T DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*T)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q3Z
 
 | |
8Q3Y
 
 | |
6FTI
 
 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Braunger, K, Becker, T, Beckmann, R. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
6FTJ
 
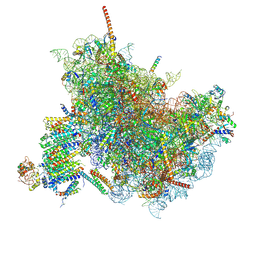 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Braunger, K, Becker, T, Beckmann, R. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
7UIN
 
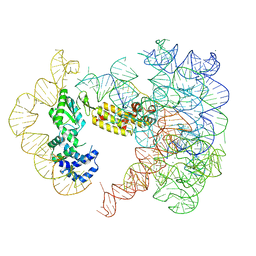 | | CryoEM Structure of an Group II Intron Retroelement | | Descriptor: | AMMONIUM ION, DNA (37-MER), E.r IIC Intron, ... | | Authors: | Chung, K, Xu, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a mobile intron retroelement poised to attack its structured DNA target
Science, 378, 2022
|
|
7UIM
 
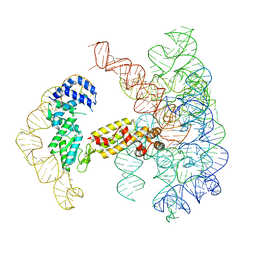 | |
9G9E
 
 | | CryoEM structure of Enterococcus italicus Csm-crRNA complex bound to AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Jungfer, K, Jinek, M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.
Nucleic Acids Res., 53, 2025
|
|
9G9H
 
 | |
9G9I
 
 | |
9G9J
 
 | |
9G9G
 
 | |
9G9B
 
 | | CryoEM structure of Enterococcus italicus Csm-crRNA (4.3) complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Jungfer, K, Jinek, M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.
Nucleic Acids Res., 53, 2025
|
|
9G9A
 
 | | CryoEM structure of Enterococcus italicus Csm-crRNA (3.2 complex) | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Jungfer, K, Jinek, M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.
Nucleic Acids Res., 53, 2025
|
|
9G9K
 
 | |
9G9F
 
 | | CryoEM structure of Enterococcus italicus Csm-crRNA-CTR complex bound to AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Jungfer, K, Jinek, M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.
Nucleic Acids Res., 53, 2025
|
|
9G9D
 
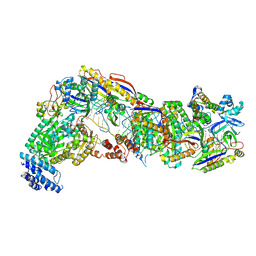 | |
9G9C
 
 | |
2JZY
 
 | |
9MQS
 
 | | CryoEM Structure of the Candida albicans Group I Intron-GMP Complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ... | | Authors: | Chung, K, Xu, L, Liu, T, Pyle, A. | | Deposit date: | 2025-01-06 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into de novo small-molecule recognition by an intron RNA structure.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9MQU
 
 | | CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Calcium Condition | | Descriptor: | CALCIUM ION, N~4~-(2-aminoethyl)-N~4~-methylpyrimidine-2,4-diamine, POTASSIUM ION, ... | | Authors: | Chung, K, Xu, L, Liu, T, Pyle, A. | | Deposit date: | 2025-01-06 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Molecular insights into de novo small-molecule recognition by an intron RNA structure.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
