4E1O
 
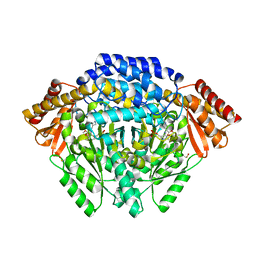 | | Human histidine decarboxylase complex with Histidine methyl ester (HME) | | Descriptor: | HISTIDINE-METHYL-ESTER, Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Komori, H, Nitta, Y, Ueno, H, Higuchi, Y. | | Deposit date: | 2012-03-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
J.Biol.Chem., 287, 2012
|
|
7L1S
 
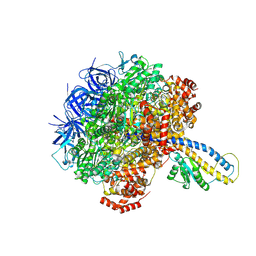 | | PS3 F1-ATPase Pi-bound Dwell | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1Q
 
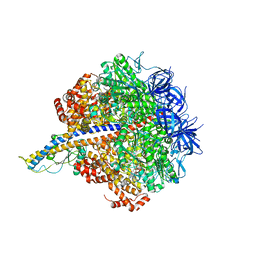 | | PS3 F1-ATPase Binding/TS Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1R
 
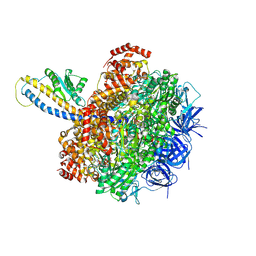 | | PS3 F1-ATPase Hydrolysis Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
5KBY
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with SYR-472 | | Descriptor: | 2-[[6-[(3~{R})-3-azanylpiperidin-1-yl]-3-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]methyl]-4-fluoranyl-benzenecarboni trile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skene, R.J, Jennings, A.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Trelagliptin (SYR-472, Zafatek), Novel Once-Weekly Treatment for Type 2 Diabetes, Inhibits Dipeptidyl Peptidase-4 (DPP-4) via a Non-Covalent Mechanism.
Plos One, 11, 2016
|
|
8SPW
 
 | | PS3 F1 Rotorless, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8SPV
 
 | | PS3 F1 Rotorless, no ATP | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8SPX
 
 | | PS3 F1 Rotorless, high ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
1DAA
 
 | |
7Y5B
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5C
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5A
 
 | | Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Wong, C.F, Saw, W.-G, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5D
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
5U7I
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 4-[3-(4-methoxyphenoxy)azetidin-1-yl]-1-methyl-3-(2-methylpropyl)-1H-pyrazolo[3,4-d]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
5U7D
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 2-(3,4-dimethoxybenzyl)-7-[(2R,3R)-2-hydroxy-6-phenylhexan-3-yl]-5-methylimidazo[5,1-f][1,2,4]triazin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
5U7K
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 3-[5-(4-ethylphenyl)-1-methyl-1H-pyrazol-4-yl]-5-propoxy[1,2,4]triazolo[4,3-a]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
4YVF
 
 | | Structure of S-adenosyl-L-homocysteine hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{[5-chloro-2-(4-chlorophenoxy)phenyl](2-{[2-(methylamino)ethyl]amino}-2-oxoethyl)amino}-N-(1,3-dihydro-2H-isoindol-2-yl)-N-methylacetamide, Adenosylhomocysteinase | | Authors: | Akiko, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and structural analyses of S-adenosyl-L-homocysteine hydrolase inhibitors based on non-adenosine analogs.
Bioorg.Med.Chem., 23, 2015
|
|
5U7L
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | (3R)-1-{3-[5-(4-ethylphenyl)-1-methyl-1H-pyrazol-4-yl]-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl}-N,N-dimethylpyrrolidin-3-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
5U7J
 
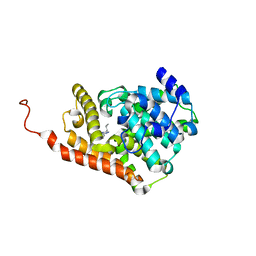 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 5-[2-(2-methoxyphenyl)ethoxy]-3-(2-methylpropyl)[1,2,4]triazolo[4,3-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
3ARN
 
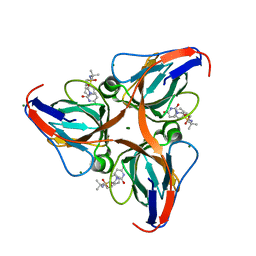 | | Human dUTPase in complex with novel uracil derivative | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, N-{5-[(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)methoxy]-2-methylpentan-2-yl}benzenesulfonamide | | Authors: | Chong, K.T, Miyahara, S, Miyakoshi, H, Fukuoka, M. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a novel class of potent human deoxyuridine triphosphatase inhibitors remarkably enhancing the antitumor activity of thymidylate synthase inhibitors
J.Med.Chem., 55, 2012
|
|
3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | Descriptor: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
7VBU
 
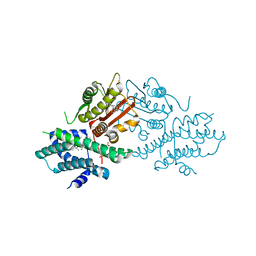 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 5 | | Descriptor: | 8-cyclopropyl-2-methyl-9H-pyrido[2,3-b]indole, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
7VBX
 
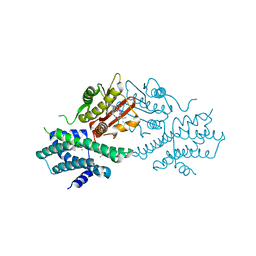 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 20 | | Descriptor: | (3~{S})-3-[5-(8-cyclopropyl-2-methyl-9~{H}-pyrido[2,3-b]indol-3-yl)-1,3,4-oxadiazol-2-yl]-4-methyl-~{N}-[(1~{R})-1-phenylethyl]pentanamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
7VBV
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 7 | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
