1LDM
 
 | |
2BK6
 
 | | The X-ray crystal structure of the Listeria innocua H31G Dps mutant. | | Descriptor: | NON-HEME IRON-CONTAINING FERRITIN | | Authors: | Ilari, A, Latella, M.C, Ribacchi, F, Su, M, Giangiacomo, L, Stefanini, S, Chasteen, N.D, Chiancone, E. | | Deposit date: | 2005-02-11 | | Release date: | 2005-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The unusual intersubunit ferroxidase center of Listeria innocua Dps is required for hydrogen peroxide detoxification but not for iron uptake. A study with site-specific mutants.
Biochemistry, 44, 2005
|
|
2BKC
 
 | |
2BJY
 
 | |
1AVT
 
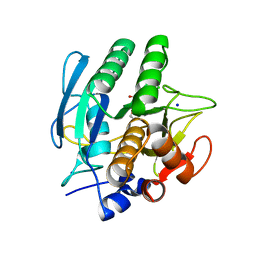 | | SUBTILISIN CARLSBERG D-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-19 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1RVE
 
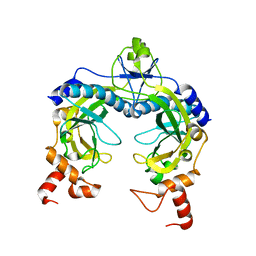 | |
1AV7
 
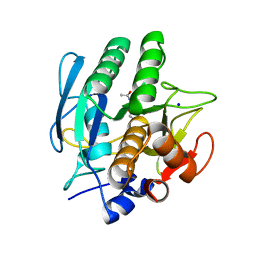 | | SUBTILISIN CARLSBERG L-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-29 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
3G4N
 
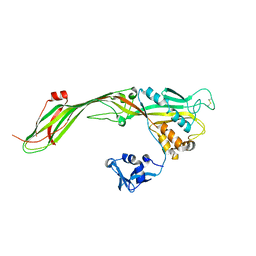 | |
3G4O
 
 | |
2GE5
 
 | |
1YO7
 
 | |
1JIG
 
 | | Dlp-2 from Bacillus anthracis | | Descriptor: | Dlp-2, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
1JI4
 
 | | NAP protein from helicobacter pylori | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, NEUTROPHIL-ACTIVATING PROTEIN A, ... | | Authors: | Zanotti, G, Papinutto, E, Dundon, W.G, Battistutta, R, Seveso, M, Del Giudice, G, Rappuoli, R, Montecucco, C. | | Deposit date: | 2001-06-29 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Neutrophil-activating Protein from Helicobacter pylori
J.Mol.Biol., 323, 2002
|
|
1JI5
 
 | | Dlp-1 from bacillus anthracis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dlp-1, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
1CZN
 
 | | REFINED STRUCTURES OF OXIDIZED FLAVODOXIN FROM ANACYSTIS NIDULANS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Smith, W.W, Pattridge, K.A, Luschinsky, C.L, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
1SBC
 
 | |
1AZ3
 
 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
1AZ4
 
 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B, T93A MUTANT | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
1C3L
 
 | | SUBTILISIN-CARLSBERG COMPLEXED WITH XENON (8 BAR) | | Descriptor: | CALCIUM ION, FORMIC ACID, SUBTILISIN-CARLSBERG, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1AZ0
 
 | | ECORV ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3'), PROTEIN (TYPE II RESTRICTION ENZYME ECORV) | | Authors: | Perona, J.J, Martin, A.M. | | Deposit date: | 1997-11-24 | | Release date: | 1998-06-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
1RV5
 
 | | COMPLEX OF ECORV ENDONUCLEASE WITH D(AAAGAT)/D(ATCTT) | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*T*AP*TP*CP*TP*T)-3', ECORV ENDONUCLEASE | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 1998-06-01 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT).
J.Mol.Biol., 277, 1998
|
|
1OFV
 
 | | FLAVODOXIN FROM ANACYSTIS NIDULANS: REFINEMENT OF TWO FORMS OF THE OXIDIZED PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Smith, W.W, Pattridge, K.A, Luschinsky, C.L, Ludwig, M.L. | | Deposit date: | 1992-06-22 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
1OPF
 
 | |
1O9R
 
 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
1PAZ
 
 | |
