2A0T
 
 | | NMR structure of the FHA1 domain of Rad53 in complex with a biological relevant phosphopeptide derived from Madt1 | | Descriptor: | Hypothetical 73.8 kDa protein in SAS3-SEC17 intergenic region, residues 301-310, Serine/threonine-protein kinase RAD53 | | Authors: | Mahajan, A, Yuan, C, Pike, B.L, Heierhorst, J, Chang, C.-F, Tsai, M.-D. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | FHA Domain-Ligand Interactions: Importance of Integrating Chemical and Biological Approaches
J.Am.Chem.Soc., 127, 2005
|
|
2WDW
 
 | | The Native Crystal Structure of the Primary Hexose Oxidase (Dbv29) in Antibiotic A40926 Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PUTATIVE HEXOSE OXIDASE | | Authors: | Liu, Y.-C, Li, Y.-S, Lyu, S.-Y, Chen, Y.-H, Chan, H.-C, Huang, C.-J, Huang, Y.-T, Chen, G.-H, Chou, C.-C, Tsai, M.-D, Li, T.-L. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Interception of Teicoplanin Oxidation Intermediates Yields New Antimicrobial Scaffolds
Nat.Chem.Biol., 7, 2011
|
|
1CEH
 
 | | STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Kumar, A, Sekharudu, C, Ramakrishnan, B, Dupureur, C.M, Zhu, H, Tsai, M.-D, Sundaralingam, M. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water.
Protein Sci., 3, 1994
|
|
1FHR
 
 | | SOLUTION STRUCTURE OF THE FHA2 DOMAIN OF RAD53 COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Byeon, I.-J.L, Liao, H, Yongkiettrakul, S, Tsai, M.-D. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides.
J.Mol.Biol., 302, 2000
|
|
1FHQ
 
 | |
1DC2
 
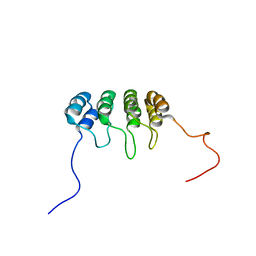 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
1T6M
 
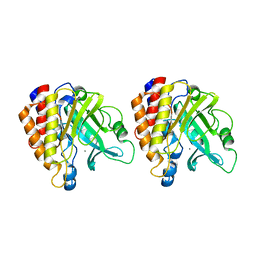 | | X-ray Structure of the R70D PI-PLC enzyme: Insight into the role of calcium and surrounding amino acids on active site geometry and catalysis. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, CALCIUM ION | | Authors: | Apiyo, D, Zhao, L, Tsai, M.-D, Selby, T.L. | | Deposit date: | 2004-05-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | X-ray Structure of the R69D Phosphatidylinositol-Specific Phospholipase C Enzyme: Insight into the Role of Calcium and Surrounding Amino Acids in Active Site Geometry and Catalysis.
Biochemistry, 44, 2005
|
|
1FDK
 
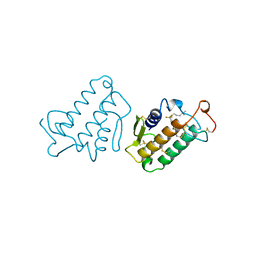 | | CARBOXYLIC ESTER HYDROLASE (PLA2-MJ33 INHIBITOR COMPLEX) | | Descriptor: | 1-DECYL-3-TRIFLUORO ETHYL-SN-GLYCERO-2-PHOSPHOMETHANOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-09-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the complex of bovine pancreatic phospholipase A2 with the inhibitor 1-hexadecyl-3-(trifluoroethyl)-sn-glycero-2-phosphomethanol,.
Biochemistry, 36, 1997
|
|
1UNE
 
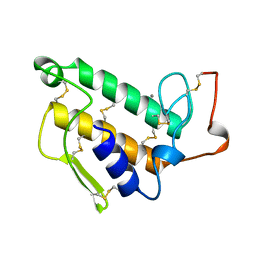 | |
1QU5
 
 | |
1HUO
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1HUZ
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1MKV
 
 | |
1MKT
 
 | |
2AFF
 
 | | The solution structure of the Ki67FHA/hNIFK(226-269)3P complex | | Descriptor: | Antigen KI-67, MKI67 FHA domain interacting nucleolar phosphoprotein | | Authors: | Byeon, I.-J.L, Li, H, Song, H, Gronenborn, A.M, Tsai, M.D. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
1J4K
 
 | |
1J4L
 
 | |
1K2N
 
 | |
1K2M
 
 | |
5ZM0
 
 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Gusti Ngurah Putu, E.P, Maestre-Reyna, M, Tsai, M.-D, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-03-31 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii
Nucleic Acids Res., 46, 2018
|
|
1J4Q
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T192) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1J4P
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3N
 
 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
