7S42
 
 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-acetamido-3,6-dideoxy-D-galactose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S3W
 
 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S44
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S43
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S45
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Acetyl Coenzyme A and dTDP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7TXS
 
 | | X-ray structure of the VioB N-aetyltransferase from Acinetobacter baumannii in the presence of a reaction intermediate | | Descriptor: | SODIUM ION, VioB, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (3R)-4-({3-[(2-{[(1S)-1-{[(2R,3S,4S,5R,6R)-4,5-dihydroxy-6-{[(R)-hydroxy{[(R)-hydroxy{[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)oxolan-2-yl]methoxy}phosphoryl]oxy}phosphoryl]oxy}-2-methyloxan-3-yl]amino}ethyl]sulfanyl}ethyl)amino]-3-oxopropyl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate (non-preferred name) | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7TXP
 
 | | X-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in complex with TDP-4-amino-4,6-dideoxy-D-glucose | | Descriptor: | SODIUM ION, VioB, dTDP-4-amino-4,6-dideoxyglucose | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7TXQ
 
 | | x-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in the present of TDP and Acetyl-CoenzymeA | | Descriptor: | ACETYL COENZYME *A, SODIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
2AQO
 
 | | Crystal structure of E. coli Isoaspartyl Dipeptidase Mutant E77Q | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
2AQV
 
 | | Crystal Structure of E. coli Isoaspartyl Dipeptidase mutant Y137F | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
6NBP
 
 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
5BJV
 
 | | X-ray structure of the PglF UDP-N-acetylglucosamine 4,6-dehydratase from Campylobacterjejuni, D396N/K397A variant in complex with UDP-N-acrtylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJU
 
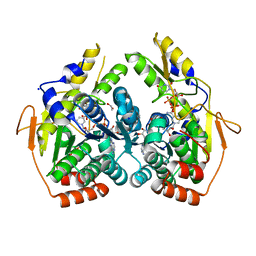 | | X-ray structure of the PglF dehydratase from Campylobacter jejuni in complex with UDP and NAD(H) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJW
 
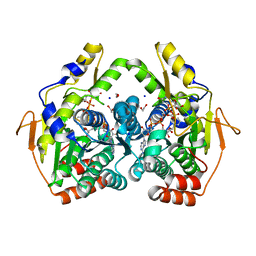 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, T595S variant, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BUV
 
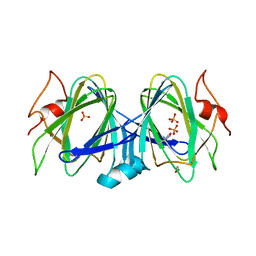 | | X-ray structure of WbcA from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, PHOSPHATE ION, ... | | Authors: | Holden, H.M, Salinger, A.J, Brown, H.A, Thoden, J.B. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical studies on WbcA, a sugar epimerase from Yersinia enterocolitica.
Protein Sci., 24, 2015
|
|
5BJX
 
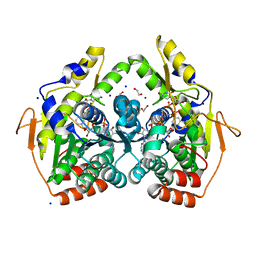 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, variant T395V, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJY
 
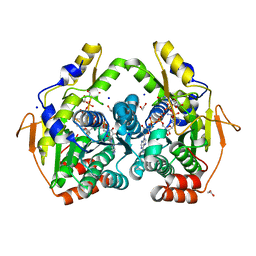 | | x-ray structure of the PglF 4,5-dehydratase from campylobacter jejuni, variant M405Y, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
6OFU
 
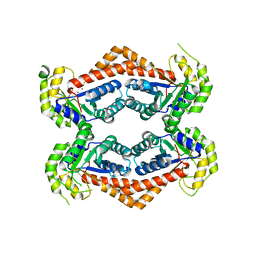 | | X-ray crystal structure of the YdjI aldolase from Escherichia coli K12 | | Descriptor: | CHLORIDE ION, YdjI aldolase, ZINC ION | | Authors: | Dopkins, B.J, Thoden, J.B, Huddleston, J.P, Narindoshvili, T, Fose, B, Rachel, F.M, Holden, H.M. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Characterization of YdjI, an Aldolase of Unknown Specificity inEscherichia coliK12.
Biochemistry, 58, 2019
|
|
6Q03
 
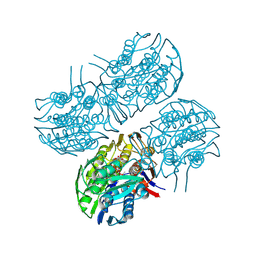 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
6Q0A
 
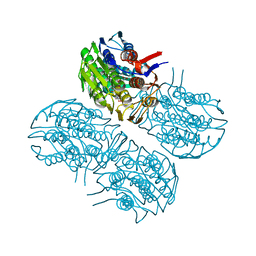 | | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid
To Be Published
|
|
6Q11
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID
To Be Published
|
|
6PZ2
 
 | |
7MFO
 
 | | X-ray structure of the L136 Aminotransferase from Acanthamoeba polyphaga mimivirus in the presence of TDP and PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFP
 
 | | crystal structure of the L136 aminotransferase K185A from acanthamoeba polyphaga mimivirus in the presence of the UDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-dihydroxy-5-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyloxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFQ
 
 | | crystal structure of the L136 aminotransferase from acanthamoeba polyphaga mimivirus in complex with the TDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
