4V6V
 
 | | Tetracycline resistance protein Tet(O) bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Atkinson, G.C, Thakor, N.S, Allas, U, Lu, C, Chan, K.Y, Tenson, T, Schulten, K, Wilson, K.S, Hauryliuk, V, Frank, J. | | Deposit date: | 2013-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Mechanism of tetracycline resistance by ribosomal protection protein Tet(O).
Nat Commun, 4, 2013
|
|
6ENJ
 
 | |
6ENU
 
 | | Polyproline-stalled ribosome in the presence of elongation-factor P (EF-P) | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-22 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
6ENF
 
 | | Cryo-EM structure of a polyproline-stalled ribosome in the absence of EF-P | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-04 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
8AP4
 
 | |
7NRD
 
 | | Structure of the yeast Gcn1 bound to a colliding stalled 80S ribosome with MBF1, A/P-tRNA and P/E-tRNA | | Descriptor: | 25S rRNA (3184-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NRC
 
 | | Structure of the yeast Gcn1 bound to a leading stalled 80S ribosome with Rbg2, Gir2, A- and P-tRNA and eIF5A | | Descriptor: | 18S rRNA (1771-MER), 25S rRNA (3184-MER), 40S ribosomal protein S0-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6EX0
 
 | |
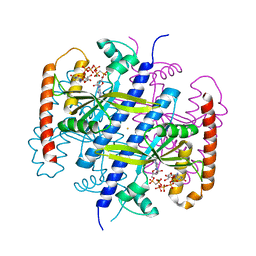
6EWZ
 
 | |
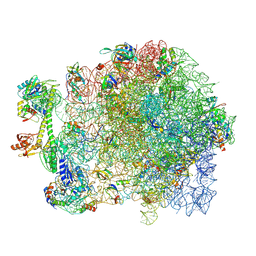
7AS9
 
 | |
7AS8
 
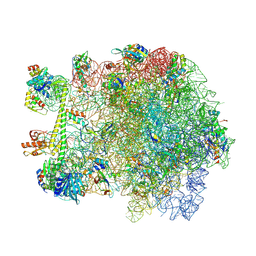 | | Bacillus subtilis ribosome quality control complex state B. Ribosomal 50S subunit with P-tRNA, RqcH, and RqcP/YabO | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for Bacterial Ribosome-Associated Quality Control by RqcH and RqcP.
Mol.Cell, 81, 2021
|
|
7ASA
 
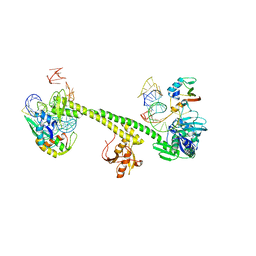 | | Bacillus subtilis ribosome-associated quality control complex state B, multibody refinement focussed on RqcH. Ribosomal 50S subunit with P-tRNA, RqcH, and RqcP/YabO | | Descriptor: | 23S rRNA, 50S ribosomal protein L11, Rqc2 homolog RqcH, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Bacterial Ribosome-Associated Quality Control by RqcH and RqcP.
Mol.Cell, 81, 2021
|
|
8S1U
 
 | | YlmH bound to stalled 50S subunits with RqcH and PtRNA | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Paternoga, H, Wilson, D.N. | | Deposit date: | 2024-02-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.
Nucleic Acids Res., 52, 2024
|
|
8S1P
 
 | | YlmH bound to PtRNA-50S | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2024-02-15 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.
Nucleic Acids Res., 52, 2024
|
|
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDU
 
 | | Rnase R bound to a 30S degradation intermediate (main state) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CED
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDV
 
 | | Rnase R bound to a 30S degradation intermediate (state II) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
