1UMZ
 
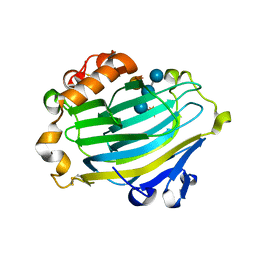 | | Xyloglucan endotransglycosylase in complex with the xyloglucan nonasaccharide XLLG. | | Descriptor: | XYLOGLUCAN ENDOTRANSGLYCOSYLASE, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A.M, Henriksson, H, Denman, S.E, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1UN1
 
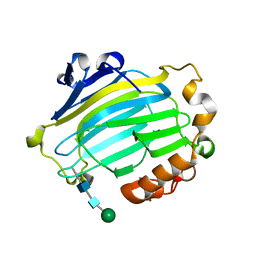 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
2UWB
 
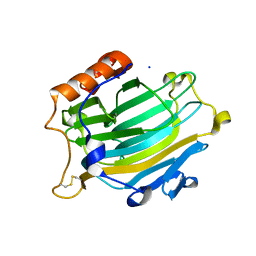 | | Crystal structure of the Nasturtium seedling mutant xyloglucanase isoform NXG1-delta-YNIIG | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2UWC
 
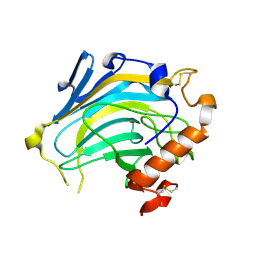 | | Crystal structure of Nasturtium xyloglucan hydrolase isoform NXG2 | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J.M, Michel, G, Kallas, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2UWA
 
 | | Crystal structure of the Nasturtium seedling xyloglucanase isoform NXG1 | | Descriptor: | CELLULASE, GLYCEROL | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-19 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
5CEL
 
 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
6CEL
 
 | | CBH1 (E212Q) CELLOPENTAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
7CEL
 
 | | CBH1 (E217Q) IN COMPLEX WITH CELLOHEXAOSE AND CELLOBIOSE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
3CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
2CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1EGN
 
 | | CELLOBIOHYDROLASE CEL7A (E223S, A224H, L225V, T226A, D262G) MUTANT | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE CEL7A, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2000-02-16 | | Release date: | 2001-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/ A224H/L225V/T226A/D262G mutant.
Biochem.J., 356, 2001
|
|
1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
1Q2E
 
 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1QJW
 
 | | CEL6A (Y169F) WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
4CEL
 
 | | ACTIVE-SITE MUTANT D214N DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1CB2
 
 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | Authors: | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | Deposit date: | 1995-11-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
1CEL
 
 | |
1QK0
 
 | | CEL6A WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-IODO-BENZYL ALCOHOL, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-08 | | Release date: | 1999-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1QK2
 
 | | WILD TYPE CEL6A WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside, ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-09 | | Release date: | 1999-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1EG1
 
 | | ENDOGLUCANASE I FROM TRICHODERMA REESEI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Kleywegt, G.J, Zou, J.-Y, Jones, T.A. | | Deposit date: | 1996-11-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 A resolution, and a comparison with related enzymes.
J.Mol.Biol., 272, 1997
|
|
1HGW
 
 | | CEL6A D175A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), COBALT (II) ION, ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
1HGY
 
 | | CEL6A D221A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose, ... | | Authors: | Zou, J.-Y, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
1DY4
 
 | | CBH1 IN COMPLEX WITH S-PROPRANOLOL | | Descriptor: | 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Stahlberg, J, Henriksson, H, Divne, C, Isaksson, R, Pettersson, G, Johansson, G, Jones, T.A. | | Deposit date: | 2000-01-26 | | Release date: | 2000-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution
J.Mol.Biol., 305, 2001
|
|
2VH9
 
 | | CRYSTAL STRUCTURE OF NXG1-DELTAYNIIG IN COMPLEX WITH XLLG, A XYLOGLUCAN DERIVED OLIGOSACCHARIDE | | Descriptor: | CELLULASE, GLYCEROL, ZINC ION, ... | | Authors: | Czjzek, M, Mark, P, Baumann, M.J, Eklof, J.M, Michel, G, Brumer, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of Nasturtium Tmnxg1 Complexes by Crystallography and Molecular Dynamics Provides Detailed Insight Into Substrate Recognition by Family Gh16 Xyloglucan Endo-Transglycosylases and Endo-Hydrolases.
Proteins, 75, 2009
|
|
