3GCC
 
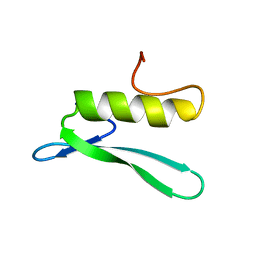 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1GCC
 
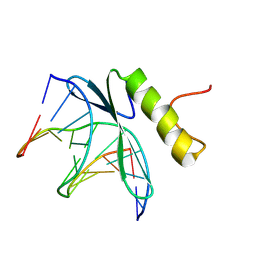 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF GCC-BOX BINDING DOMAIN OF ATERF1 AND GCC-BOX DNA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*GP*CP*GP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*CP*GP*CP*CP*AP*GP*C)-3'), ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 | | Authors: | Yamasaki, K, Allen, M.D, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1ILE
 
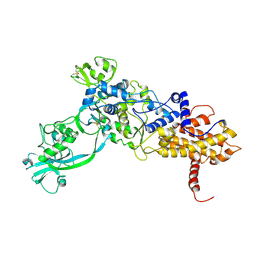 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
2GCC
 
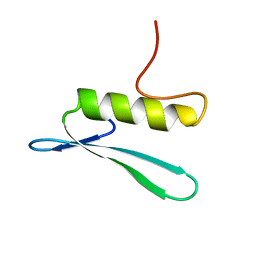 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1RRB
 
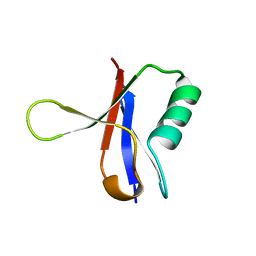 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVB
 
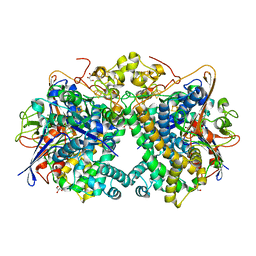 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVC
 
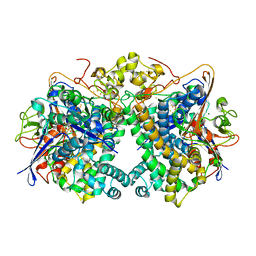 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
6A87
 
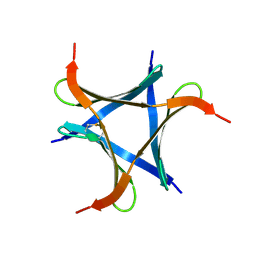 | | Pholiota squarrosa lectin (PhoSL) in complex with fucose(alpha1-6)GlcNAc | | Descriptor: | METHANETHIOL, alpha-L-fucopyranose, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamasaki, K, Yamasaki, T, Kubota, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for specific recognition of core fucosylation in N-glycans by Pholiota squarrosa lectin (PhoSL).
Glycobiology, 29, 2019
|
|
6A86
 
 | | Pholiota squarrosa lectin | | Descriptor: | (3R)-butane-1,3-diol, lectin | | Authors: | Yamasaki, K, Yamasaki, T, Kubota, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific recognition of core fucosylation in N-glycans by Pholiota squarrosa lectin (PhoSL).
Glycobiology, 29, 2019
|
|
1A8H
 
 | | METHIONYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sugiura, I, Nureki, O, Ugaji, Y, Kuwabara, S, Lober, B, Giege, R, Moras, D, Yokoyama, S, Konno, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
Structure, 8, 2000
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
1UL5
 
 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 7 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 7 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1UL4
 
 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 4 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 4 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1WJ2
 
 | | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | | Descriptor: | Probable WRKY transcription factor 4, ZINC ION | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis WRKY DNA binding domain.
Plant Cell, 17, 2005
|
|
1WID
 
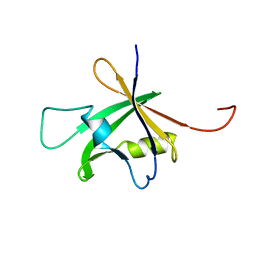 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
1YSE
 
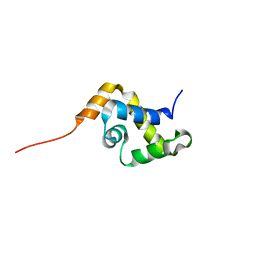 | | Solution structure of the MAR-binding domain of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Yamasaki, K, Yamaguchi, H. | | Deposit date: | 2005-02-08 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Mode of the Matrix Attachment Region-binding Domain of the Transcription Factor SATB1 That Regulates the T-cell Maturation
J.Biol.Chem., 281, 2006
|
|
3VU8
 
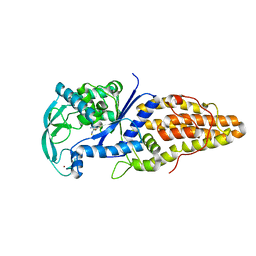 | | Metionyl-tRNA synthetase from Thermus thermophilus complexed with methionyl-adenylate analogue | | Descriptor: | Methionine--tRNA ligase, N-[METHIONYL]-N'-[ADENOSYL]-DIAMINOSULFONE, ZINC ION | | Authors: | Konno, M, Kato-Murayama, M, Toma-Fukai, S, Uchikawa, E, Nureki, O, Yokoyama, S. | | Deposit date: | 2012-06-22 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The modeling of structures of specific conformation of homosysteine-AMP leading to thiolactone-formation on class Ia aminoacyl-tRNA synthetases
To be Published
|
|
