5AY4
 
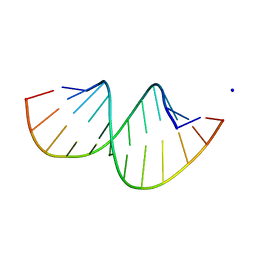 | | Crystal structure of RNA duplex containing C-C base pairs obtained in the presence of Hg(II) | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*UP*(CBR)P*GP*AP*CP*UP*CP*C)-3'), SODIUM ION | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Saneyoshi, H, Okamoto, I, Tanaka, Y, Ono, A. | | Deposit date: | 2015-08-06 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of a Silver(I)-RNA Hybrid Duplex Containing Watson-Crick-like CSilver(I)C Metallo-Base Pairs
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AY2
 
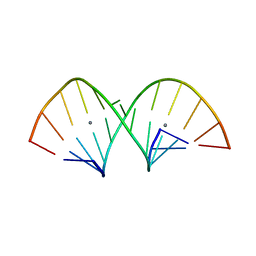 | | Crystal structure of RNA duplex containing C-Ag(I)-C base pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*UP*(CBR)P*GP*AP*CP*UP*CP*C)-3'), SILVER ION | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Saneyoshi, H, Okamoto, I, Tanaka, Y, Ono, A. | | Deposit date: | 2015-08-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Crystal Structure of a Silver(I)-RNA Hybrid Duplex Containing Watson-Crick-like CSilver(I)C Metallo-Base Pairs
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AY3
 
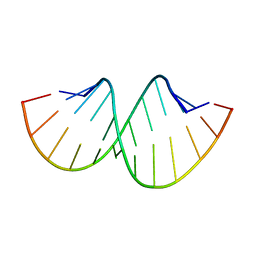 | | Crystal structure of RNA duplex containing C-C base pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*UP*(CBR)P*GP*A*CP*UP*CP*C)-3') | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Saneyoshi, H, Okamoto, I, Tanaka, Y, Ono, A. | | Deposit date: | 2015-08-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-Resolution Crystal Structure of a Silver(I)-RNA Hybrid Duplex Containing Watson-Crick-like CSilver(I)C Metallo-Base Pairs
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5EXV
 
 | |
8J4C
 
 | | YeeE(TsuA)-YeeD(TsuB) complex for thiosulfate uptake | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Spirochaeta thermophila YeeE(TsuA)-YeeD(TsuB),UPF0033 domain-containing protein, SirA-like domain-containing protein (chimera), ... | | Authors: | Ikei, M, Miyazaki, R, Monden, K, Naito, Y, Takeuchi, A, Takahashi, Y.S, Tanaka, Y, Ichikawa, M, Tsukazaki, T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structure and function of YeeE-YeeD complex for sophisticated thiosulfate uptake
To Be Published
|
|
1UD9
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
4G5H
 
 | | Crystal structure of capsular polysaccharide synthesizing enzyme CapE from Staphylococcus aureus in complex with by-product | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap8E, FORMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Miyafusa, T, Caaveiro, J.M.M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-08-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dynamic elements govern the catalytic activity of CapE, a capsular polysaccharide-synthesizing enzyme from Staphylococcus aureus.
Febs Lett., 587, 2013
|
|
1WSC
 
 | | Crystal structure of ST0229, function unknown protein from Sulfolobus tokodaii | | Descriptor: | Hypothetical protein ST0229 | | Authors: | Murayama, T, Tanaka, Y, Sasaki, T, Yasutake, Y, Yao, M, Tsumoto, K, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of ST0229, function unknown protein from Sulfolobus tokodaii
To be Published
|
|
1WOZ
 
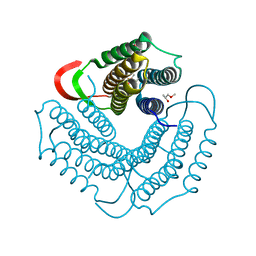 | | Crystal structure of uncharacterized protein ST1454 from Sulfolobus tokodaii | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 177aa long conserved hypothetical protein (ST1454) | | Authors: | Sasaki, T, Tanaka, Y, Yasutake, Y, Yao, M, Tanaka, I, Tsumoto, K, Kumagai, I. | | Deposit date: | 2004-08-27 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the uncharacterized protein ST1454 from Sulfolobus tokodaii.
To be Published
|
|
2ZKL
 
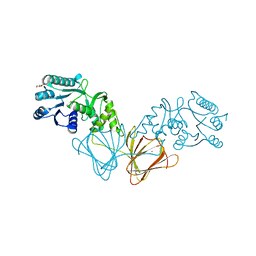 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Yao, M, Tanaka, I, Tsumoto, K. | | Deposit date: | 2008-03-25 | | Release date: | 2009-03-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of capsular polysaccharide assembling protein from Staphylococcus aureus
to be published
|
|
5B4E
 
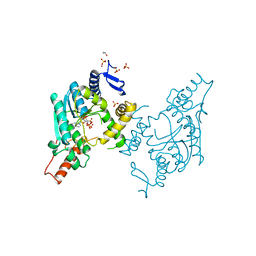 | | Sulfur Transferase TtuA in complex with iron sulfur cluster and ATP derivative | | Descriptor: | IRON/SULFUR CLUSTER, ISOPROPYL ALCOHOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5B4F
 
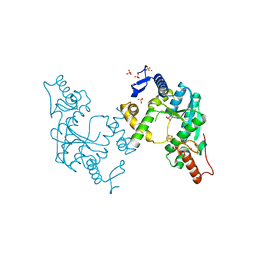 | | Sulfur Transferase TtuA in complex with iron sulfur cluster | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, Sulfur Transferase TtuA, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DCV
 
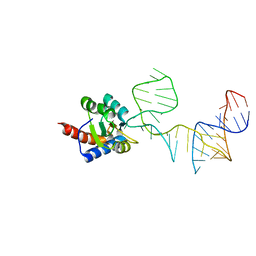 | | Crystal structure of PhoRpp38-SL12M complex | | Descriptor: | 50S ribosomal protein L7Ae, RNA (47-MER) | | Authors: | Oshima, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis for recognition of a kink-turn motif by an archaeal homologue of human RNase P protein Rpp38
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5H6O
 
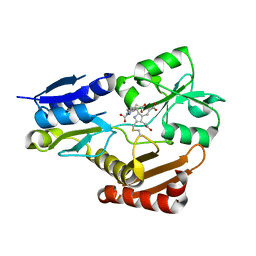 | | Porphobilinogen deaminase from Vibrio Cholerae | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, MAGNESIUM ION, Porphobilinogen deaminase | | Authors: | Funamizu, T, Chen, M, Tanaka, Y, Ishimori, K, Uchida, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Porphobilinogen deaminase from Vibrio Cholerae
To Be Published
|
|
8K1R
 
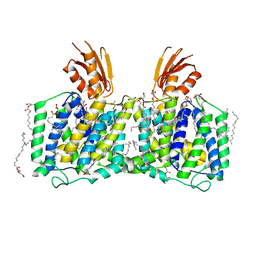 | | YeeE(TsuA)-YeeD(TsuB) complex for thiosulfate uptake | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Spirochaeta thermophila YeeE(TsuA)-YeeD(TsuB),UPF0033 domain-containing protein, SirA-like domain-containing protein (chimera) | | Authors: | Ikei, M, Miyazaki, R, Monden, K, Naito, Y, Takeuchi, A, Takahashi, Y.S, Tanaka, Y, Ichikawa, M, Tsukazaki, T. | | Deposit date: | 2023-07-11 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of YeeE-YeeD complex for sophisticated thiosulfate uptake
To Be Published
|
|
3B0V
 
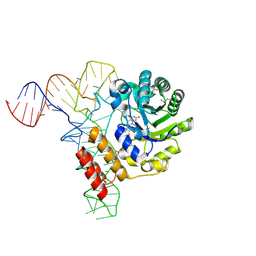 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0U
 
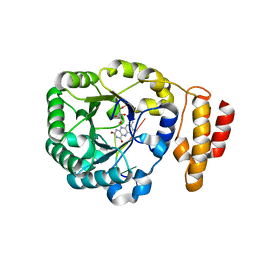 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA fragment | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*(H2U)P*A)-3'), tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0P
 
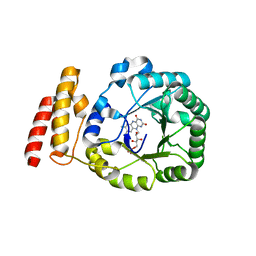 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AZV
 
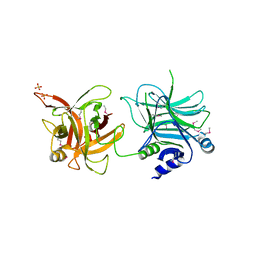 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
3AZW
 
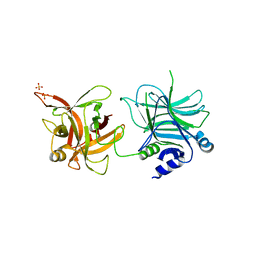 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
8IFC
 
 | | Arbekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 2024
|
|
8IFE
 
 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 2024
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 2024
|
|
8IFB
 
 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 2024
|
|
