3WON
 
 | | Crystal structure of the DAP BII dipeptide complex III | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOM
 
 | | Crystal structure of the DAP BII dipeptide complex II | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOO
 
 | | Crystal structure of the DAP BII hexapeptide complex I | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOI
 
 | | Crystal structure of the DAP BII (S657A) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOK
 
 | | Crystal structure of the DAP BII (Space) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOJ
 
 | | Crystal structure of the DAP BII | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3VP8
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p | | Descriptor: | General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
2DLC
 
 | | Crystal structure of the ternary complex of yeast tyrosyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, O-(ADENOSINE-5'-O-YL)-N-(L-TYROSYL)PHOSPHORAMIDATE, T-RNA (76-MER), ... | | Authors: | Tsunoda, M, Kusakabe, Y, Tanaka, N, Nakamura, K.T. | | Deposit date: | 2006-04-18 | | Release date: | 2007-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
Nucleic Acids Res., 35, 2007
|
|
3ANX
 
 | | Crystal structure of triamine/agmatine aminopropyltransferase (SPEE) from thermus thermophilus, complexed with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, spermidine synthase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Tanaka, N, Oshima, T, Kumasaka, T. | | Deposit date: | 2010-09-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
3AUA
 
 | | Crystal structure of the quaternary complex-2 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3AU8
 
 | | Crystal structure of the ternary complex of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3AU9
 
 | | Crystal structure of the quaternary complex-1 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
2D2X
 
 | | Crystal structure of 2-deoxy-scyllo-inosose synthase | | Descriptor: | 2-deoxy-scyllo-inosose synthase, COBALT (II) ION, GLYCEROL, ... | | Authors: | Nango, E, Kumasaka, T, Tanaka, N, Kakinuma, K, Eguchi, T. | | Deposit date: | 2005-09-20 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+
Proteins, 70, 2008
|
|
2DCJ
 
 | | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2DCK
 
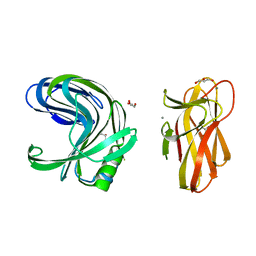 | | A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | CALCIUM ION, GLYCEROL, xylanase J | | Authors: | Fibriansah, G, Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
3WA6
 
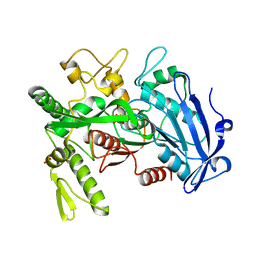 | |
3WA7
 
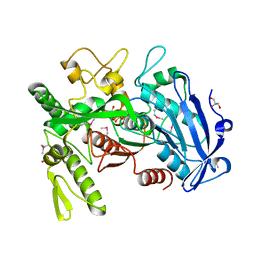 | | Crystal structure of selenomethionine-labeled tannase from Lactobacillus plantarum in the orthorhombic crystal | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Matoba, Y, Tanaka, N, Sugiyama, M. | | Deposit date: | 2013-04-27 | | Release date: | 2013-07-24 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational analyses of tannase from Lactobacillus plantarum.
Proteins, 81, 2013
|
|
4FXC
 
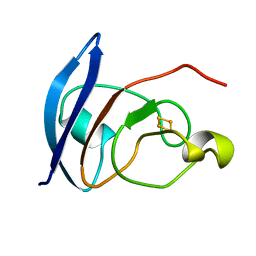 | |
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | Descriptor: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | Authors: | Fibriansah, G, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
1SRD
 
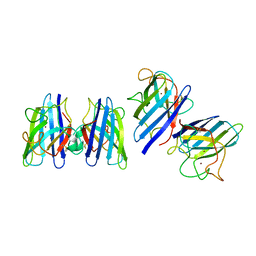 | | Three-dimensional structure of CU,ZN-superoxide dismutase from spinach at 2.0 Angstroms resolution | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Kitagawa, Y, Katsube, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Cu,Zn-superoxide dismutase from spinach at 2.0 A resolution.
J.Biochem.(Tokyo), 109, 1991
|
|
4K1O
 
 | |
4K1N
 
 | |
7ZOP
 
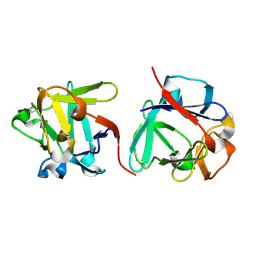 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | Descriptor: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
