1MNM
 
 | |
6D8I
 
 | |
6D8J
 
 | |
8JP6
 
 | |
8JPG
 
 | | Cryo-EM structure of full-length ERGIC-53 with MCFD2 | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53, ... | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | Structure of full-length ERGIC-53 in complex with MCFD2 for cargo transport.
Nat Commun, 15, 2024
|
|
8JP8
 
 | |
8JP4
 
 | |
8JP7
 
 | |
8JP9
 
 | |
8JP5
 
 | |
7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | Authors: | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
5GU6
 
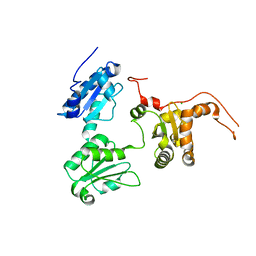 | | Crystal structure of Human ERp44 form I | | Descriptor: | CHLORIDE ION, Endoplasmic reticulum resident protein 44 | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1YTF
 
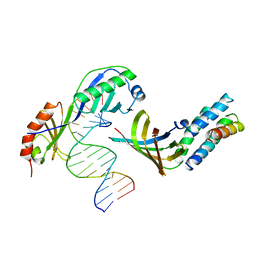 | | YEAST TFIIA/TBP/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*AP*TP*AP*TP*AP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*TP*AP*TP*AP*TP*AP*AP*AP*AP*C)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)), ... | | Authors: | Tan, S, Hunziker, Y, Sargent, D.F, Richmond, T.J. | | Deposit date: | 1996-04-05 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a yeast TFIIA/TBP/DNA complex.
Nature, 381, 1996
|
|
8WT4
 
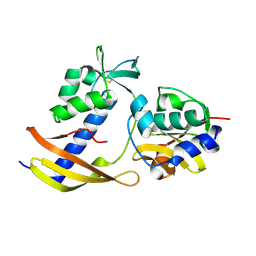 | |
5XJ4
 
 | | Complex structure of durvalumab-scFv/PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab-VH, durvalumab-VL | | Authors: | Tan, S, Liu, K, Chai, Y, Gao, G.F, Qi, J. | | Deposit date: | 2017-04-29 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab
Protein Cell, 9, 2018
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
3IA0
 
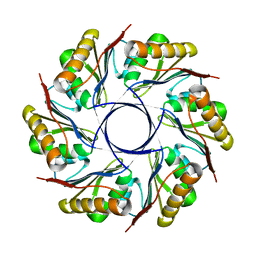 | |
3I96
 
 | | Ethanolamine Utilization Microcompartment Shell Subunit, EutS | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Ethanolamine utilization protein eutS, ... | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2009-07-10 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanisms of a Protein-Based Organelle in Escherichia coli.
Science, 327, 2010
|
|
3I87
 
 | |
3I71
 
 | |
3I82
 
 | |
6SYM
 
 | |
3I6P
 
 | |
5GU7
 
 | | Crystal Structure of human ERp44 form II | | Descriptor: | Endoplasmic reticulum resident protein 44 | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7OT4
 
 | |
