1O5P
 
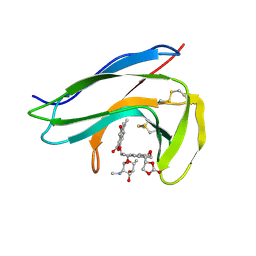 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
2D0S
 
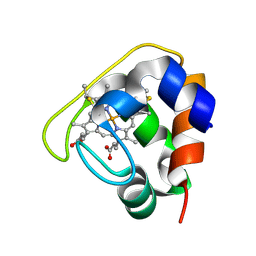 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
2AI5
 
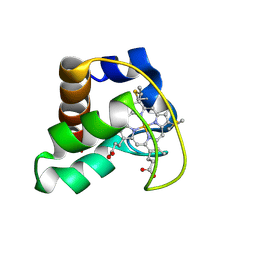 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-07-29 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
7DEN
 
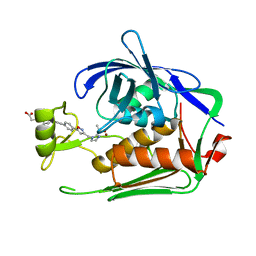 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-[(1~{R},5~{S})-6-[2-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]ethynyl]-3-azabicyclo[3.1.0]hexan-3-yl]butanoic acid, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Takashima, H. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7CID
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 1-[3-(4-chlorophenyl)propyl]imidazole, DIMETHYL SULFOXIDE, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIB
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-oxidanyl-4-phenyl-benzoic acid, DIMETHYL SULFOXIDE, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI7
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R,3R)-2-azanyl-1-[4-[[4-[2-[4-(hydroxymethyl)phenyl]ethynyl]phenyl]methyl]piperidin-1-yl]-4-methylsulfonyl-3-oxidanyl-butan-1-one, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI6
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[3-(4-chlorophenyl)propyl]imidazol-2-yl]ethanol, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI8
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[(5-phenyl-1,2-oxazol-3-yl)methyl]imidazol-2-yl]ethanol, MAGNESIUM ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIE
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-3-oxidanyl-N-[3-(trifluoromethyloxy)phenyl]propanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIA
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI4
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-4-methylsulfonyl-N-[3-(trifluoromethyloxy)phenyl]butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIC
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-azanyl-N-[3-(trifluoromethyloxy)phenyl]ethanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI9
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-2-[[4-[2-[3-[[2-[(1S)-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenoxy]methyl]propane-1,3-diol, PHOSPHATE ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI5
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (3R)-3-azanyl-4-oxidanylidene-4-[[3-(trifluoromethyloxy)phenyl]amino]butanoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7DEL
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 3-[4-[2-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenyl]propan-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Tanaka-Yamamoto, N. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7DEM
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 5-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]pent-4-yn-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Matsuda, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
