1GVJ
 
 | |
1XGN
 
 | |
1XGM
 
 | |
1XGO
 
 | |
2E42
 
 | | Crystal structure of C/EBPbeta Bzip homodimer V285A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2E43
 
 | | Crystal structure of C/EBPbeta Bzip homodimer K269A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
1XGS
 
 | |
1USO
 
 | |
1USM
 
 | |
7N2M
 
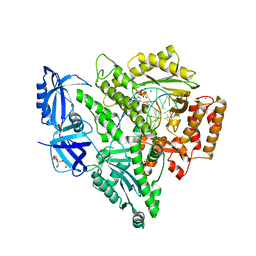 | | Crystal structure of DNA polymerase alpha catalytic core in complex with dCTP and template/primer having T-C mismatch at the post-insertion site | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*T*AP*GP*TP*CP*GP*CP*TP*CP*CP*AP*GP*GP*C)-3'), DNA polymerase alpha catalytic subunit, ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2021-05-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into mismatch extension by human DNA polymerase alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3JTG
 
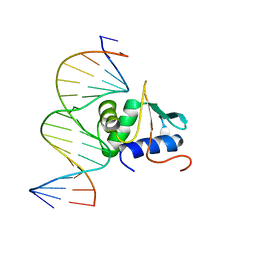 | | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*CP*AP*GP*GP*AP*AP*AP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*GP*TP*TP*TP*CP*CP*TP*GP*TP*TP*T)-3'), ETS-related transcription factor Elf-3 | | Authors: | Tahirov, T.H, Babayeva, N.D, Agarkar, V.B, Rizzino, A. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA.
J.Mol.Biol., 397, 2010
|
|
6AS7
 
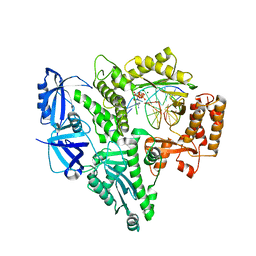 | | CRYSTAL STRUCTURE OF THE CATALYTIC CORE OF HUMAN DNA POLYMERASE ALPHA IN TERNARY COMPLEX WITH AN DNA-PRIMED DNA TEMPLATE AND DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, COBALT (II) ION, DNA (5'-D(*AP*GP*GP*CP*GP*CP*TP*CP*CP*AP*GP*GP*C)-3'), ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Activity and fidelity of human DNA polymerase alpha depend on primer structure.
J. Biol. Chem., 293, 2018
|
|
5EXR
 
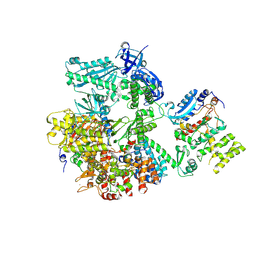 | | Crystal structure of human primosome | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2015-11-24 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome.
J.Biol.Chem., 291, 2016
|
|
5F0S
 
 | | Crystal structure of C-terminal domain of the human DNA primase large subunit with bound DNA template/RNA primer and manganese ion | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*CP*AP*AP*CP*AP*TP*A)-3'), DNA primase large subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2015-11-28 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome.
J.Biol.Chem., 291, 2016
|
|
5F0Q
 
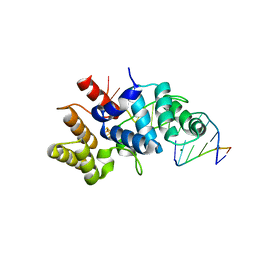 | | Crystal structure of C-terminal domain of the human DNA primase large subunit with bound DNA template/RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*CP*AP*AP*CP*AP*TP*A)-3'), DNA primase large subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2015-11-28 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome.
J.Biol.Chem., 291, 2016
|
|
4L0Y
 
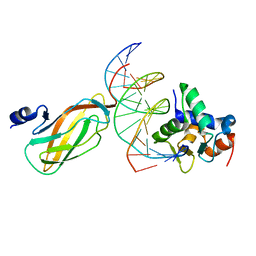 | |
4L18
 
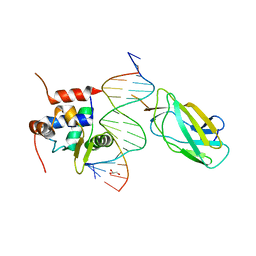 | |
4L0Z
 
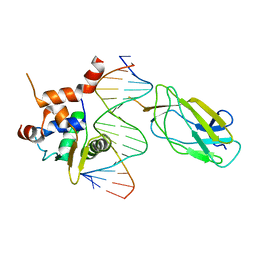 | |
2BJ3
 
 | | NIKR-apo | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICKEL RESPONSIVE REGULATOR | | Authors: | Tahirov, T.H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Pyrococcus Horikoshii Nikr: Nickel Sensing and Implications for the Regulation of DNA Recognition
J.Mol.Biol., 348, 2005
|
|
2BJ9
 
 | | NIKR with bound NICKEL and phosphate | | Descriptor: | NICKEL (II) ION, NICKEL RESPONSIVE REGULATOR, PHOSPHATE ION, ... | | Authors: | Tahirov, T.H, Chivers, P.T. | | Deposit date: | 2005-01-31 | | Release date: | 2005-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Pyrococcus Horikoshii Nikr: Nickel Sensing and Implications for the Regulation of DNA Recognition
J.Mol.Biol., 348, 2005
|
|
2BJ8
 
 | |
2BJ1
 
 | | NIKR IN OPEN CONFORMATION AND NICKEL BOUND TO HIGH-AFFINITY SITES | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, NICKEL RESPONSIVE REGULATOR | | Authors: | Tahirov, T.H. | | Deposit date: | 2005-01-27 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Pyrococcus Horikoshii Nikr: Nickel Sensing and Implications for the Regulation of DNA Recognition
J.Mol.Biol., 348, 2005
|
|
2BJ7
 
 | |
1V1A
 
 | |
1V1B
 
 | |
