6J9U
 
 | | Complex structure of Lactobacillus casei lactate dehydrogenase penta mutant with pyruvate | | Descriptor: | L-lactate dehydrogenase, PYRUVIC ACID, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
6JML
 
 | | Re-refined structure of R-state L-lactate dehydrogenase fromLactobacillus casei | | Descriptor: | L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
5XXL
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXO
 
 | | Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXM
 
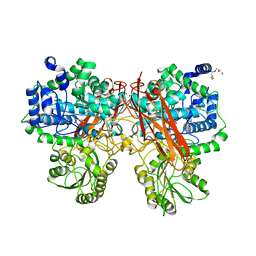 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YSD
 
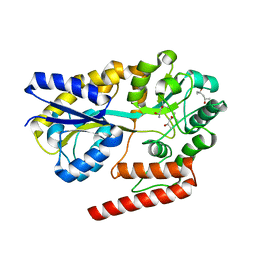 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
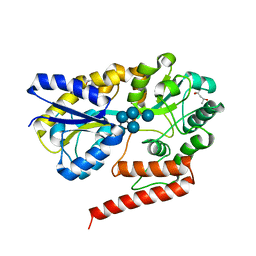 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
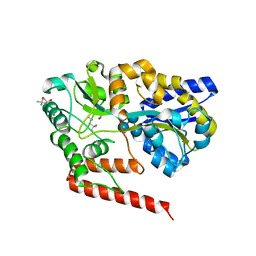 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
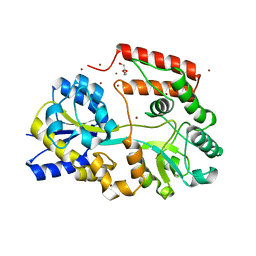 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5Z20
 
 | | The ternary structure of D-lactate dehydrogenase from Pseudomonas aeruginosa with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase (Fermentative), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
5Z21
 
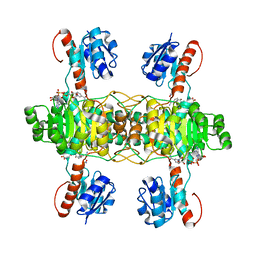 | | The ternary structure of D-lactate dehydrogenase from Fusobacterium nucleatum with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase, OXAMIC ACID | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
5Z1Z
 
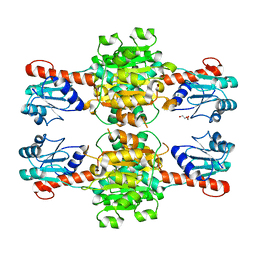 | | The apo-structure of D-lactate dehydrogenase from Escherichia coli | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
6ABI
 
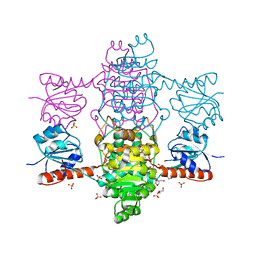 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6ABJ
 
 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
3VPH
 
 | | L-lactate dehydrogenase from Thermus caldophilus GK24 complexed with oxamate, NADH and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
3VPG
 
 | | L-lactate dehydrogenase from Thermus caldophilus GK24 | | Descriptor: | GLYCEROL, L-lactate dehydrogenase | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
3WX0
 
 | |
3WFJ
 
 | | The complex structure of D-mandelate dehydrogenase with NADH | | Descriptor: | 2-dehydropantoate 2-reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
3WFI
 
 | | The crystal structure of D-mandelate dehydrogenase | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
3WVL
 
 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
3W99
 
 | | Crystal Structure of Human Nucleosome Core Particle lacking H4 N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
3W98
 
 | | Crystal Structure of Human Nucleosome Core Particle lacking H3.1 N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
3W97
 
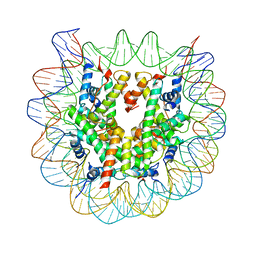 | | Crystal Structure of Human Nucleosome Core Particle lacking H2B N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
