1UEL
 
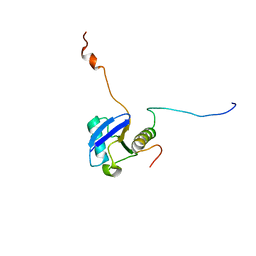 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | 著者 | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-05-19 | | 公開日 | 2004-02-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
2D07
 
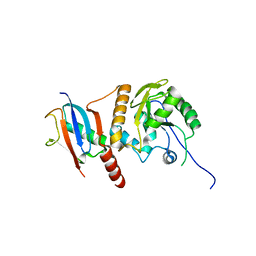 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | 分子名称: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | 著者 | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | 登録日 | 2005-07-26 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
4YM6
 
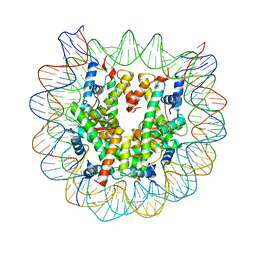 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | 分子名称: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.514 Å) | | 主引用文献 | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4YM5
 
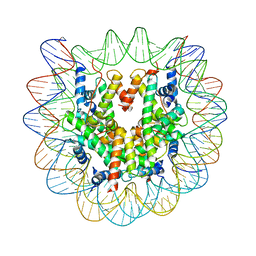 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | 分子名称: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | 著者 | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.005 Å) | | 主引用文献 | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
1WYW
 
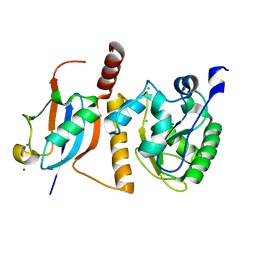 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | 分子名称: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | 著者 | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | 登録日 | 2005-02-17 | | 公開日 | 2005-06-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
5JRG
 
 | | Crystal structure of the nucleosome containing the DNA with tetrahydrofuran (THF) | | 分子名称: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | 著者 | Osakabe, A, Arimura, Y, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2016-05-06 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Polymorphism of apyrimidinic DNA structures in the nucleosome
Sci Rep, 7, 2017
|
|
4A11
 
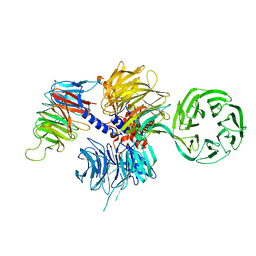 | | Structure of the hsDDB1-hsCSA complex | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | 著者 | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | 登録日 | 2011-09-13 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
5B24
 
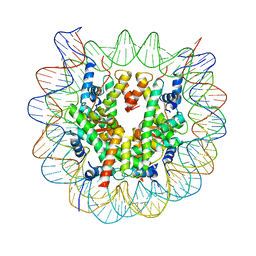 | | The crystal structure of the nucleosome containing cyclobutane pyrimidine dimer | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Horikoshi, N, Tachiwana, H, Kagawa, W, Osakabe, A, Kurumizaka, H. | | 登録日 | 2015-12-31 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structure of the nucleosome containing ultraviolet light-induced cyclobutane pyrimidine dimer
Biochem.Biophys.Res.Commun., 471, 2016
|
|
4A0K
 
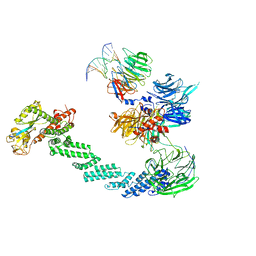 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | 分子名称: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (5.93 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
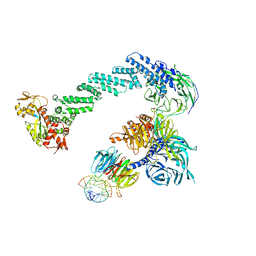 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | 分子名称: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (7.4 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
2RVB
 
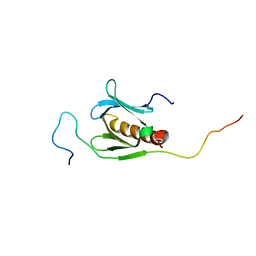 | |
4WSN
 
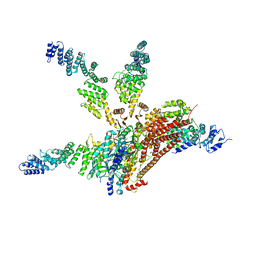 | | Crystal structure of the COP9 signalosome, a P1 crystal form | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-10-28 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome.
Nature, 531, 2016
|
|
8EBT
 
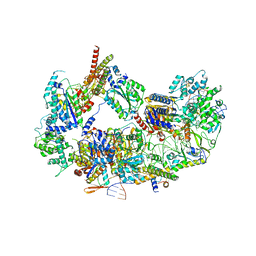 | |
8EBS
 
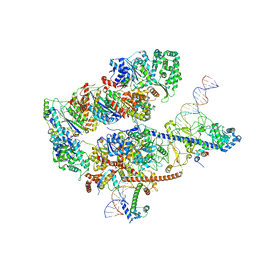 | |
8EBW
 
 | |
8EBV
 
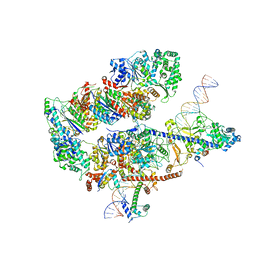 | |
8EBX
 
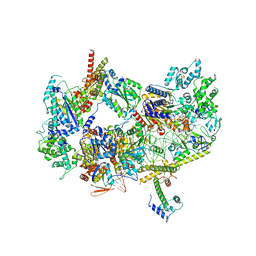 | |
8EBY
 
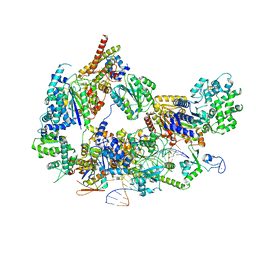 | | XPC release from Core7-XPA-DNA (AP) | | 分子名称: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | 著者 | Kim, J, Yang, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-05-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBU
 
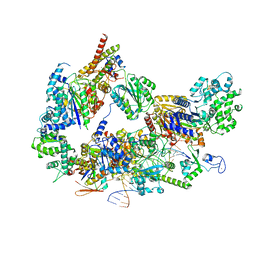 | | XPC release from Core7-XPA-DNA (Cy5) | | 分子名称: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | 著者 | Kim, J, Yang, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-05-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
6R8Z
 
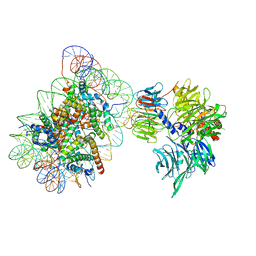 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
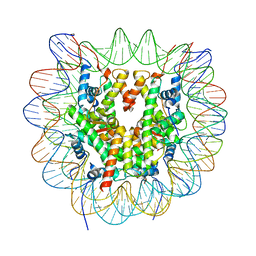 | | Cryo-EM structure of NCP_THF2(-3) | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R90
 
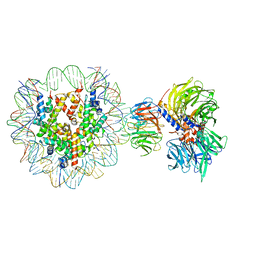 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R91
 
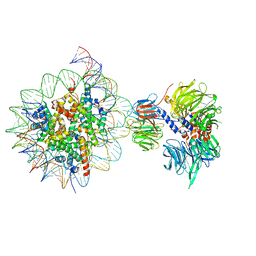 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
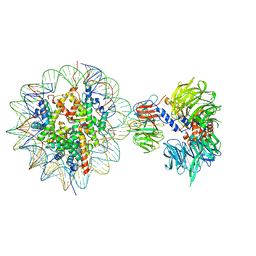 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
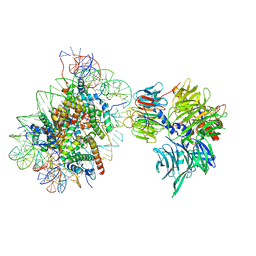 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
