4XCM
 
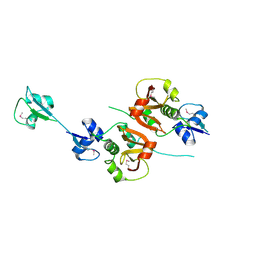 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5LS2
 
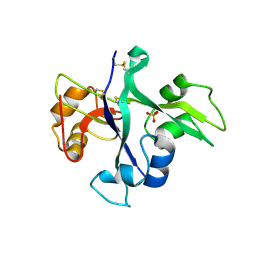 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9QRS
 
 | | Crystal structure of CERK6 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, J.X.J, Gysel, K, Stougaard, J, Andersen, K.R. | | Deposit date: | 2025-04-04 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
6QUP
 
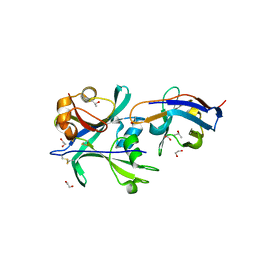 | | Structural signatures in EPR3 define a unique class of plant carbohydrate receptors | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, ... | | Authors: | Wong, J.E, Gysel, K, Birkefeldt, T.G, Vinther, M, Muszynski, A, Azadi, P, Laursen, N.S, Sullivan, J.T, Ronson, C.W, Stougaard, J, Andersen, K.R. | | Deposit date: | 2019-02-28 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Structural signatures in EPR3 define a unique class of plant carbohydrate receptors.
Nat Commun, 11, 2020
|
|
4ACJ
 
 | | Crystal structure of the TLDC domain of Oxidation resistance protein 2 from zebrafish | | Descriptor: | WU:FB25H12 PROTEIN, | | Authors: | Blaise, M, B Alsarraf, H.M.A, Wong, J.E.M.M, Midtgaard, S.R, Laroche, F, Schack, L, Spaink, H, Stougaard, J, Thirup, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of the Tldc Domain of Oxidation Resistance Protein 2 from Zebrafish.
Proteins, 80, 2012
|
|
9GB9
 
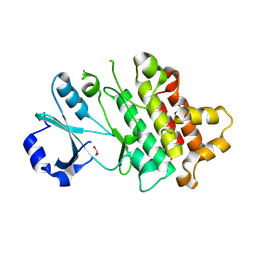 | | Crystal structure of Lotus japonicus CERK6 kinase domain D460N | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, LysM type receptor kinase | | Authors: | Simonsen, B.W, Gysel, K, Andersen, K.R. | | Deposit date: | 2024-07-30 | | Release date: | 2024-11-27 | | Last modified: | 2025-12-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two residues reprogram immunity receptors for nitrogen-fixing symbiosis.
Nature, 648, 2025
|
|
8PS7
 
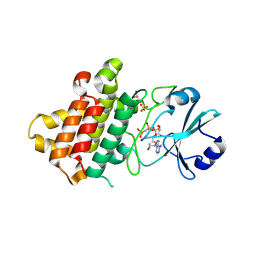 | | Crystal structure of Medicago truncatula LYR4 kinase domain | | Descriptor: | GLYCEROL, LysM-domain receptor-like kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Simonsen, B, Gysel, K, Laursen, M, Andersen, K.R. | | Deposit date: | 2023-07-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Medicago truncatula LYR4 intracellular domain serves as a scaffold in immunity signaling independent of its phosphorylation activity.
New Phytol., 246, 2025
|
|
9GFZ
 
 | | Crystal structure of Medicago Truncatula LYK3 kinase domain D459N | | Descriptor: | LysM domain receptor-like kinase 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Simonsen, B, Gysel, K, Andersen, C.G, Andersen, K.R. | | Deposit date: | 2024-08-12 | | Release date: | 2024-11-27 | | Last modified: | 2025-12-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two residues reprogram immunity receptors for nitrogen-fixing symbiosis.
Nature, 648, 2025
|
|
4UZ3
 
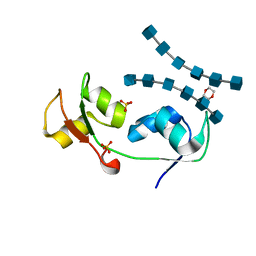 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Wong, J.E.M.M, Blaise, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UZ2
 
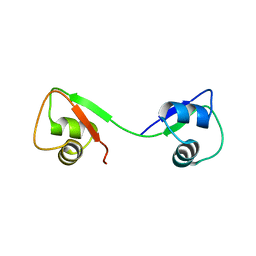 | |
6XWE
 
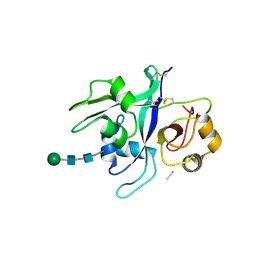 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
8S79
 
 | | Lotus japonicus NFR5 intracellular domain in complex with Nanobody 200 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hansen, S.B, Gysel, K, Andersen, K.R. | | Deposit date: | 2024-02-29 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A conserved juxtamembrane motif in plant NFR5 receptors is essential for root nodule symbiosis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7BAX
 
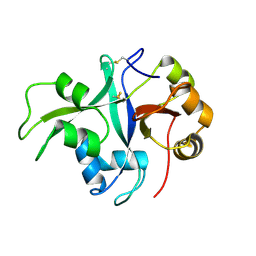 | | Crystal structure of LYS11 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase | | Authors: | Laursen, M, Cheng, J, Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-12-16 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AU7
 
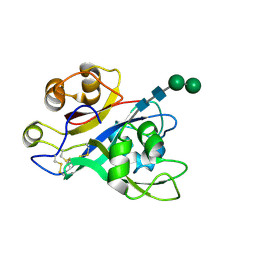 | | Crystal structure of Nod Factor Perception ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8PEH
 
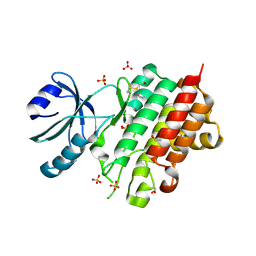 | | Crystal structure of Lotus japonicus SYMRK kinase domain D738N | | Descriptor: | 1,2-ETHANEDIOL, Receptor-like kinase SYMRK, SULFATE ION | | Authors: | Noergaard, M.M.M, Gysel, K, Hansen, S.B, Andersen, K.R. | | Deposit date: | 2023-06-14 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of the alpha-I motif in SYMRK drives root nodule organogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9Q83
 
 | | Crystal structure of Lotus japonicus CHIP13 in complex with chitooctaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Andersen, K.R. | | Deposit date: | 2025-02-21 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
9Q84
 
 | | Crystal structure of Lotus japonicus CHIP13 extracellular domain in complex with chitooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Andersen, K.R. | | Deposit date: | 2025-02-21 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
9GXZ
 
 | |
9GXF
 
 | |
9H24
 
 | |
9H39
 
 | |
9H3B
 
 | | Lotus japonicus CERK6 extracellular domain in complex with chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, ... | | Authors: | Hansen, S.B, Gysel, K, Andersen, K.R. | | Deposit date: | 2024-10-16 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
9H6V
 
 | | Lotus japonicus CERK6 extracellular domain in complex with two nanobodies | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Alsarraf, H.M.A.B, Hansen, S.B, Andersen, K.R. | | Deposit date: | 2024-10-25 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
9H3A
 
 | | Crystal structure of Lotus japonicus CHIP13 extracellular domain in complex with chitooctaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Andersen, K.R. | | Deposit date: | 2024-10-16 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
